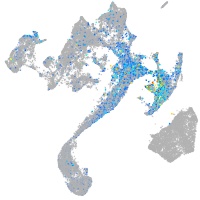
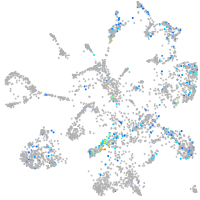
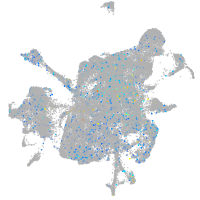
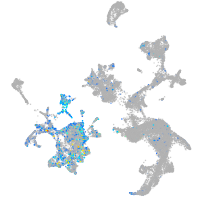
endothelin converting enzyme 1
ZFIN 
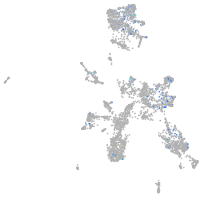
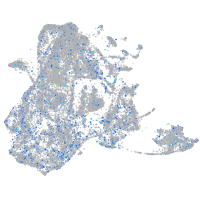
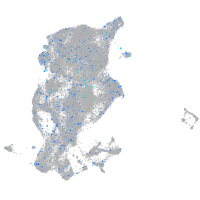
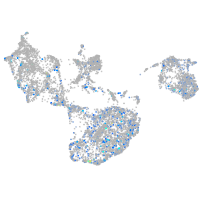
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| mnx1 | 0.757 | rps18 | -0.292 |
| nkx6.2 | 0.757 | ncl | -0.275 |
| LOC103910736 | 0.757 | pnrc2 | -0.274 |
| LOC101883836 | 0.753 | ddx5 | -0.266 |
| insm1a | 0.753 | baz1b | -0.246 |
| ins | 0.745 | ube2d2 | -0.245 |
| col9a1b | 0.742 | ppiaa | -0.236 |
| greb1l | 0.722 | rbm8a | -0.236 |
| sgpl1 | 0.722 | si:ch211-214j24.10 | -0.235 |
| myl9a | 0.721 | si:ch73-281n10.2 | -0.232 |
| atcaya | 0.707 | snrpb2 | -0.224 |
| CT573204.1 | 0.702 | akap12b | -0.221 |
| map6a | 0.699 | nop56 | -0.218 |
| rsph9 | 0.699 | sfpq | -0.213 |
| slc26a6 | 0.669 | acin1b | -0.213 |
| myl10 | 0.669 | syncrip | -0.213 |
| dhdhl | 0.664 | rhoab | -0.213 |
| pax6b | 0.663 | marcksb | -0.211 |
| gng12a | 0.655 | srsf1a | -0.210 |
| rab7b | 0.637 | top1l | -0.209 |
| parvb | 0.634 | NC-002333.4 | -0.208 |
| gng2 | 0.630 | hmgb2a | -0.198 |
| thrap3a | 0.620 | nucks1a | -0.198 |
| rbfox3b | 0.605 | nucks1b | -0.197 |
| mllt11 | 0.569 | thoc2 | -0.197 |
| neurod1 | 0.567 | tgif1 | -0.196 |
| si:ch73-25f10.6 | 0.556 | anp32e | -0.195 |
| FAM53C | 0.553 | banf1 | -0.195 |
| arfip2a | 0.551 | mki67 | -0.194 |
| myl9b | 0.535 | srsf5a | -0.194 |
| ift52 | 0.533 | zgc:110425 | -0.193 |
| st3gal7 | 0.519 | rpl6 | -0.193 |
| itgb1b.1 | 0.516 | nop58 | -0.193 |
| mtss1la | 0.512 | zmat2 | -0.193 |
| dbn1 | 0.512 | ddx4 | -0.191 |