ELL associated factor 1
ZFIN 
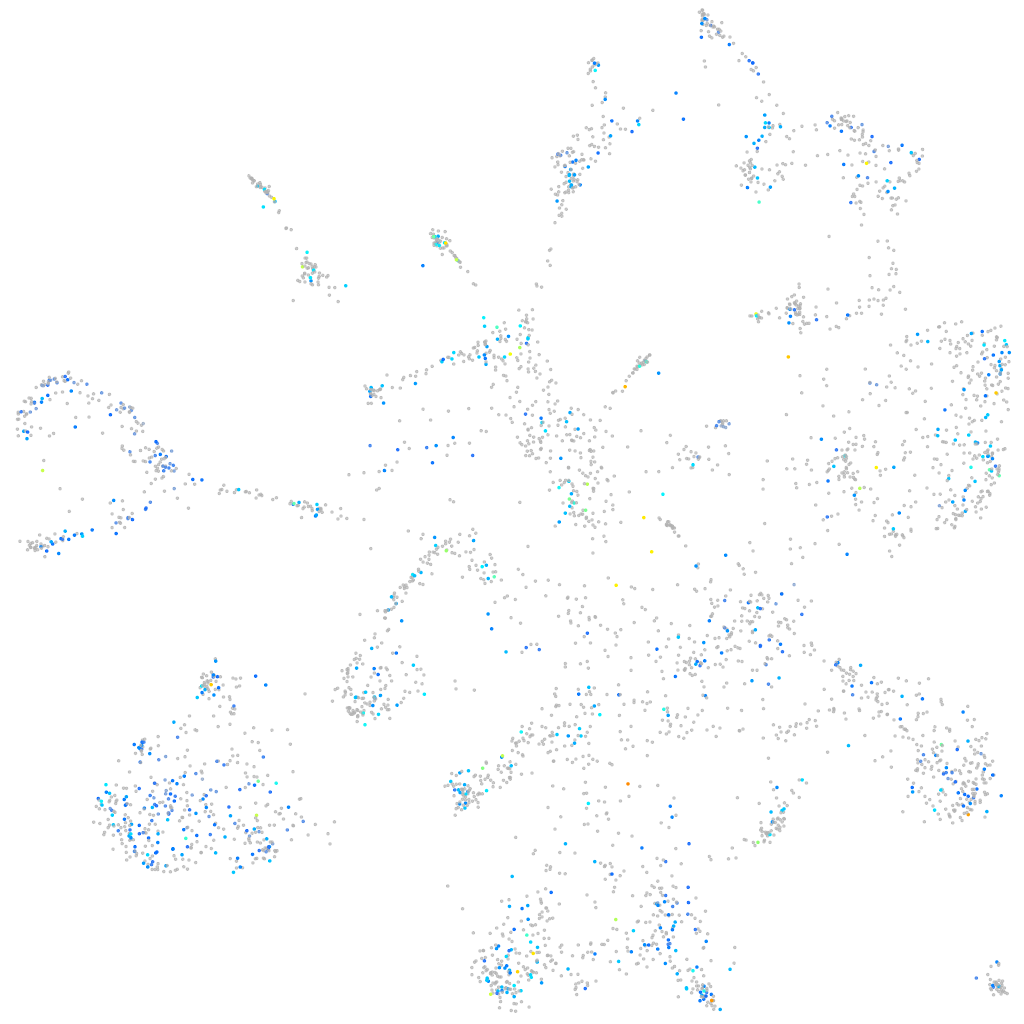
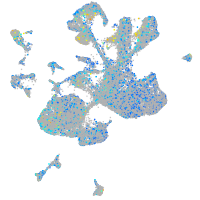
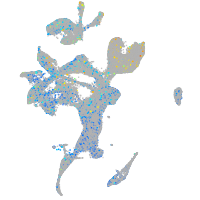
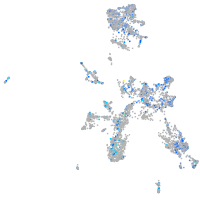
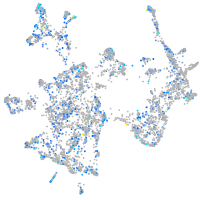
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| ppdpfb | 0.130 | rpl29 | -0.108 |
| slc25a5 | 0.114 | rps29 | -0.088 |
| pkig | 0.113 | rpl38 | -0.084 |
| gabarapb | 0.113 | rps28 | -0.074 |
| atf4b | 0.112 | NC-002333.4 | -0.073 |
| eif4a1b | 0.111 | CABZ01072614.1 | -0.072 |
| ccng1 | 0.110 | ppiaa | -0.068 |
| h2afx1 | 0.110 | LOC110439372 | -0.060 |
| moxd1l | 0.109 | hmga2 | -0.060 |
| vdac3 | 0.109 | banf1 | -0.059 |
| vdac2 | 0.109 | rpl37 | -0.058 |
| eif1b | 0.106 | rplp2l | -0.058 |
| sdcbp2 | 0.105 | si:dkey-151g10.6 | -0.057 |
| zgc:153867 | 0.104 | kif26aa | -0.056 |
| CU633823.1 | 0.101 | rpa3 | -0.054 |
| cdc42 | 0.101 | plekhg2 | -0.054 |
| mrps36 | 0.100 | fbn2b | -0.054 |
| h3f3c | 0.100 | mt-co1 | -0.053 |
| myo15ab | 0.097 | spred2b | -0.052 |
| higd2a | 0.095 | dut | -0.052 |
| GCA | 0.095 | pcdh2g17 | -0.051 |
| CT573103.1 | 0.095 | gli3 | -0.050 |
| btf3 | 0.095 | rps18 | -0.050 |
| oaz1a | 0.095 | tshz2 | -0.049 |
| ckbb | 0.095 | rpl36 | -0.049 |
| hist2h2l | 0.094 | top2a | -0.048 |
| ubc | 0.094 | rplp1 | -0.047 |
| CU655829.1 | 0.093 | pcna | -0.046 |
| eef1g | 0.092 | nradd | -0.046 |
| npdc1a | 0.092 | ndufa1 | -0.045 |
| dap1b | 0.091 | gpc4 | -0.045 |
| anxa13 | 0.091 | COX3 | -0.044 |
| tpm1 | 0.090 | rpl6 | -0.043 |
| rasgrp4 | 0.090 | fndc3bb | -0.043 |
| actb2 | 0.090 | mt-nd3 | -0.042 |