E2F transcription factor 8
ZFIN 
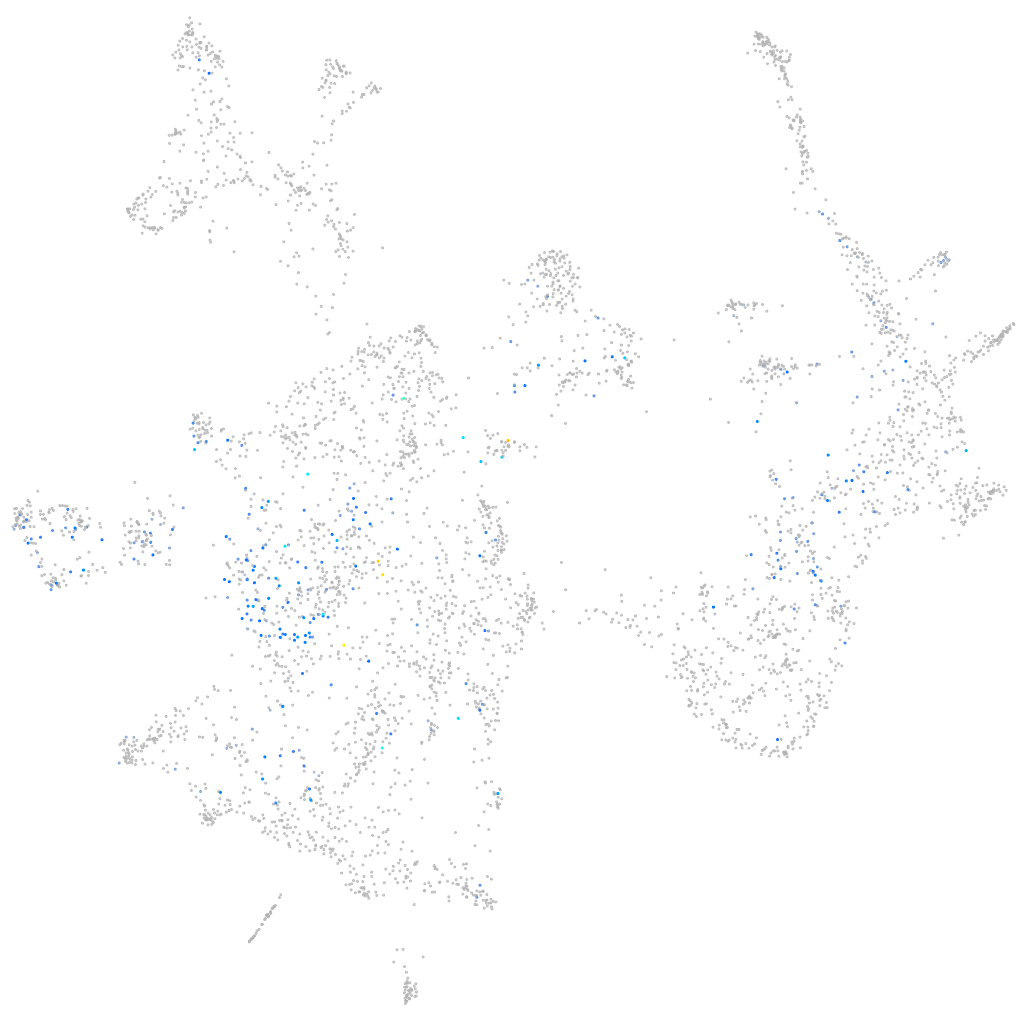
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| zgc:110540 | 0.440 | tob1b | -0.113 |
| rrm2 | 0.425 | h1f0 | -0.104 |
| CABZ01005379.1 | 0.360 | nupr1a | -0.097 |
| pcna | 0.357 | tmem59 | -0.093 |
| esco2 | 0.356 | gabarapb | -0.085 |
| asf1ba | 0.354 | atp2b1a | -0.085 |
| mibp | 0.353 | eno1a | -0.085 |
| tyms | 0.351 | ip6k2a | -0.084 |
| rrm1 | 0.350 | ppdpfb | -0.084 |
| slbp | 0.346 | gapdhs | -0.082 |
| lig1 | 0.345 | mcl1a | -0.082 |
| fbxo5 | 0.342 | wbp2nl | -0.082 |
| chaf1a | 0.334 | tspan13b | -0.081 |
| rpa2 | 0.324 | map1lc3a | -0.080 |
| dut | 0.324 | atp6v1e1b | -0.079 |
| tk1 | 0.316 | ube2h | -0.079 |
| dhfr | 0.316 | fthl27 | -0.078 |
| si:dkey-261m9.17 | 0.310 | kif1aa | -0.078 |
| cks1b | 0.308 | calm1a | -0.078 |
| prim1 | 0.302 | ppdpfa | -0.077 |
| dek | 0.294 | gabarapa | -0.077 |
| rfc4 | 0.293 | si:dkey-16p21.8 | -0.077 |
| mybl2b | 0.292 | ap3s1.1 | -0.077 |
| haus1 | 0.291 | calml4a | -0.076 |
| stmn1a | 0.291 | rnasekb | -0.076 |
| chaf1b | 0.290 | cd164l2 | -0.075 |
| rpa3 | 0.290 | adipor1a | -0.075 |
| h2afx | 0.288 | gpx2 | -0.075 |
| fen1 | 0.287 | atp1a3b | -0.075 |
| trip13 | 0.287 | otofb | -0.074 |
| slc29a2 | 0.286 | osbpl1a | -0.074 |
| cdca5 | 0.285 | itm2ba | -0.074 |
| dck | 0.280 | pkig | -0.074 |
| nsd2 | 0.279 | hist2h2l | -0.073 |
| unga | 0.277 | CR925719.1 | -0.073 |