DAZ interacting zinc finger protein 1
ZFIN 
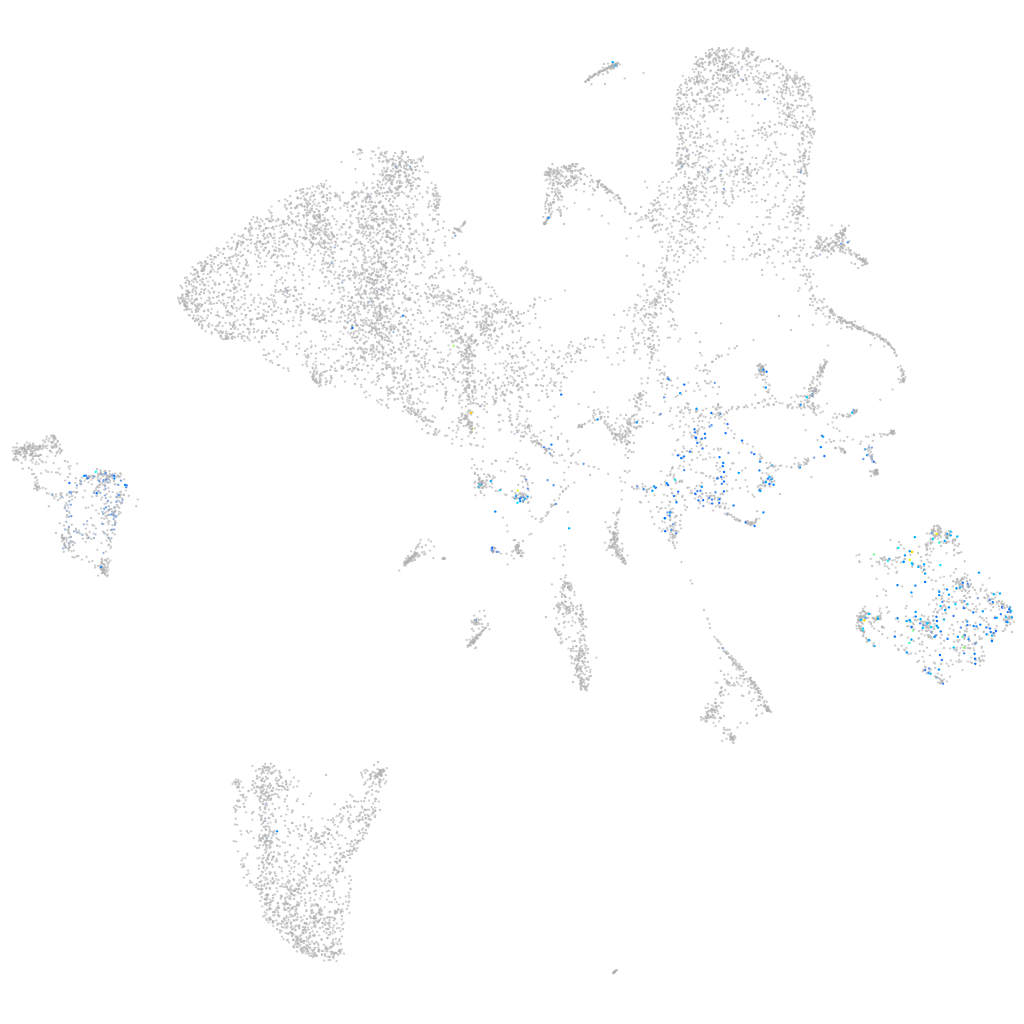
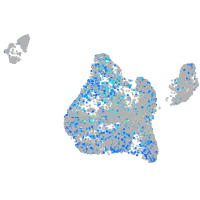
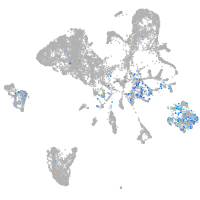
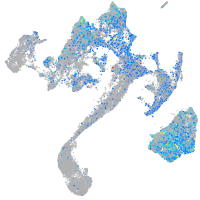
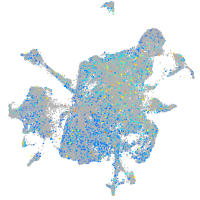
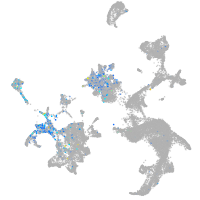
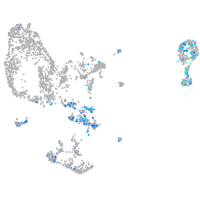
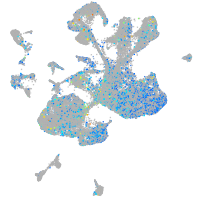
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| marcksl1b | 0.284 | rpl37 | -0.208 |
| akap12b | 0.282 | rps10 | -0.198 |
| cx43.4 | 0.281 | gapdh | -0.195 |
| marcksb | 0.278 | eno3 | -0.190 |
| hspb1 | 0.278 | ahcy | -0.189 |
| acin1a | 0.268 | zgc:114188 | -0.182 |
| si:ch211-152c2.3 | 0.261 | nme2b.1 | -0.180 |
| nucks1a | 0.255 | gamt | -0.178 |
| syncrip | 0.254 | rps17 | -0.172 |
| cdh6 | 0.252 | sod1 | -0.170 |
| smarca4a | 0.251 | atp5l | -0.166 |
| ppig | 0.244 | gatm | -0.165 |
| smc1al | 0.243 | suclg1 | -0.163 |
| ilf3b | 0.242 | atp5if1b | -0.163 |
| anp32a | 0.242 | fbp1b | -0.161 |
| hnrnpa1a | 0.241 | nupr1b | -0.159 |
| hmgb1b | 0.239 | prdx2 | -0.156 |
| snrnp70 | 0.237 | eef1da | -0.155 |
| khdrbs1a | 0.237 | dap | -0.154 |
| seta | 0.236 | gpx4a | -0.152 |
| zgc:110425 | 0.235 | gstt1a | -0.151 |
| srrm2 | 0.234 | zgc:92744 | -0.151 |
| chd7 | 0.234 | glud1b | -0.151 |
| sox11b | 0.234 | pklr | -0.150 |
| hmgb3a | 0.233 | gstp1 | -0.150 |
| hnrnph1l | 0.233 | aldob | -0.149 |
| hnrnpub | 0.233 | mdh1aa | -0.146 |
| NC-002333.4 | 0.232 | mat1a | -0.146 |
| cbx1a | 0.231 | atp5mc3b | -0.146 |
| hnrnpa1b | 0.230 | cox6b1 | -0.146 |
| qkia | 0.229 | cx32.3 | -0.145 |
| chd4b | 0.229 | atp5mc1 | -0.144 |
| hp1bp3 | 0.229 | aldh7a1 | -0.143 |
| hmga1a | 0.229 | apoa4b.1 | -0.142 |
| si:ch73-281n10.2 | 0.228 | scp2a | -0.141 |