"dynein, cytoplasmic 2, light intermediate chain 1"
ZFIN 
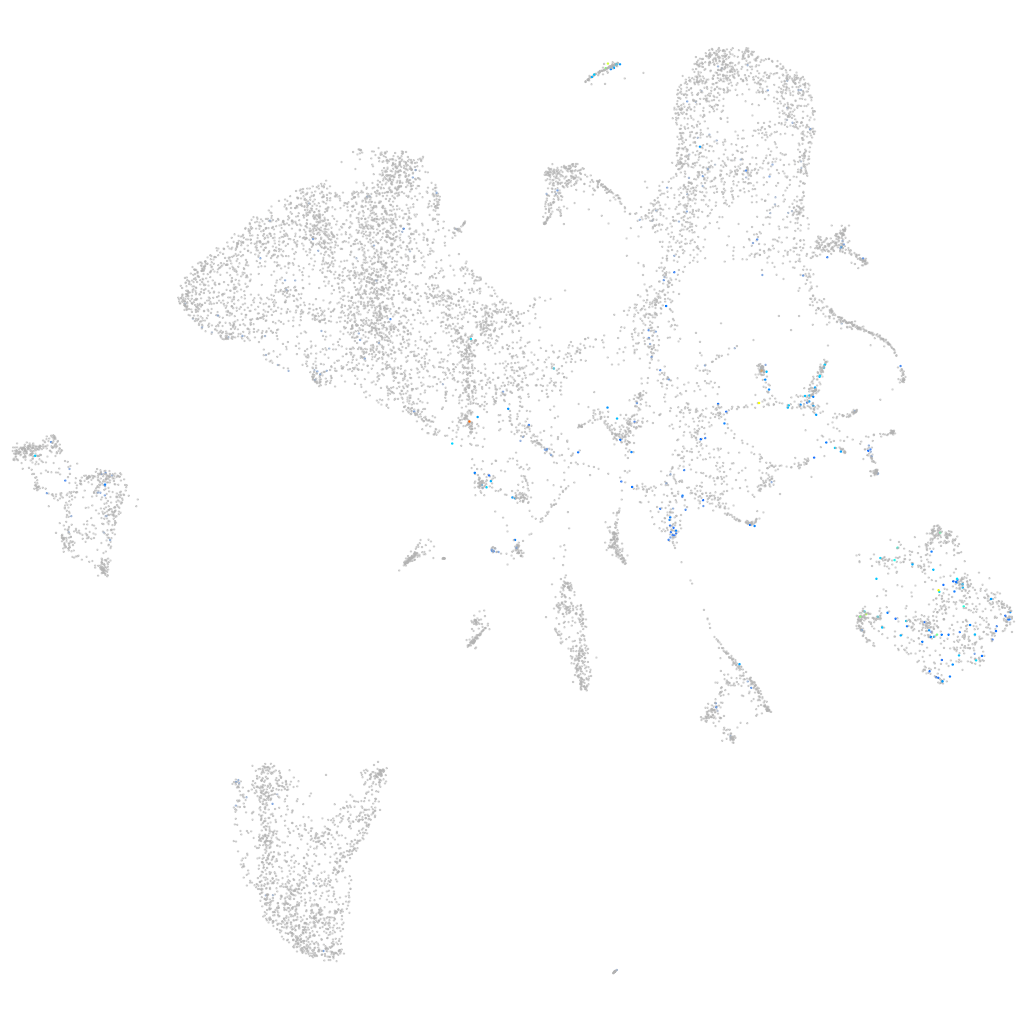
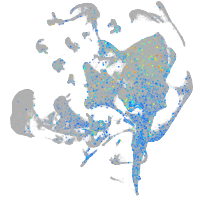
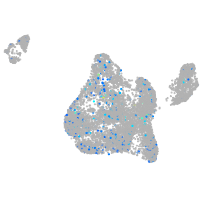
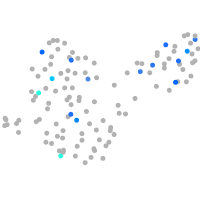
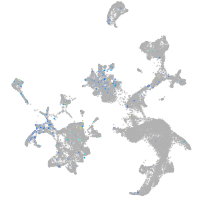
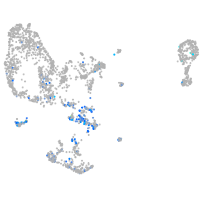
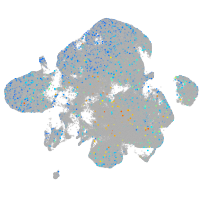
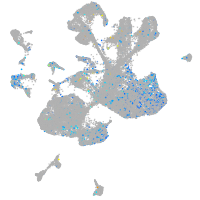
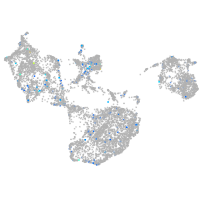
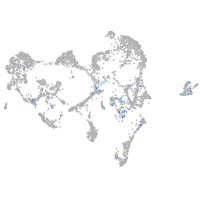
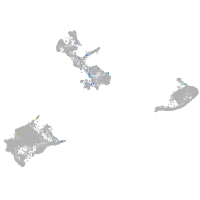
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| grid2ipa | 0.188 | ahcy | -0.116 |
| kiss1 | 0.175 | gamt | -0.115 |
| GPR139 | 0.175 | gapdh | -0.115 |
| hes6 | 0.161 | gatm | -0.103 |
| marcksl1b | 0.159 | eno3 | -0.102 |
| gabbr2 | 0.156 | nme2b.1 | -0.099 |
| CT027744.2 | 0.150 | rpl37 | -0.098 |
| si:dkey-117i10.1 | 0.149 | fbp1b | -0.096 |
| XLOC-036879 | 0.145 | bhmt | -0.094 |
| hmgn6 | 0.144 | rps10 | -0.093 |
| si:ch211-152c2.3 | 0.142 | dap | -0.093 |
| hmgb3a | 0.140 | nupr1b | -0.093 |
| nt5c1ab | 0.140 | zgc:92744 | -0.092 |
| si:ch211-194m7.3 | 0.139 | mat1a | -0.091 |
| ilf3b | 0.138 | aldh7a1 | -0.090 |
| mcoln1b | 0.136 | aldh6a1 | -0.088 |
| khdrbs1a | 0.133 | zgc:114188 | -0.088 |
| nucks1a | 0.131 | aldob | -0.088 |
| marcksb | 0.131 | ckba | -0.086 |
| dll4 | 0.130 | apoa1b | -0.086 |
| syncrip | 0.130 | glud1b | -0.085 |
| hdac1 | 0.129 | suclg1 | -0.084 |
| top1l | 0.129 | cx32.3 | -0.082 |
| tspan7 | 0.128 | afp4 | -0.082 |
| hnrnpaba | 0.128 | agxtb | -0.082 |
| akap12b | 0.128 | gstt1a | -0.082 |
| mchr2 | 0.128 | apoa4b.1 | -0.082 |
| ccl44 | 0.127 | apoc2 | -0.082 |
| tusc3 | 0.126 | scp2a | -0.082 |
| ahi1 | 0.126 | etfb | -0.081 |
| hnrnpa0a | 0.125 | gnmt | -0.081 |
| h3f3d | 0.125 | agxta | -0.080 |
| hnrnpa0b | 0.125 | aqp12 | -0.080 |
| si:ch73-1a9.3 | 0.124 | apoa2 | -0.079 |
| AL935174.5 | 0.124 | eef1da | -0.079 |