"dynein, cytoplasmic 1, light intermediate chain 1"
ZFIN 
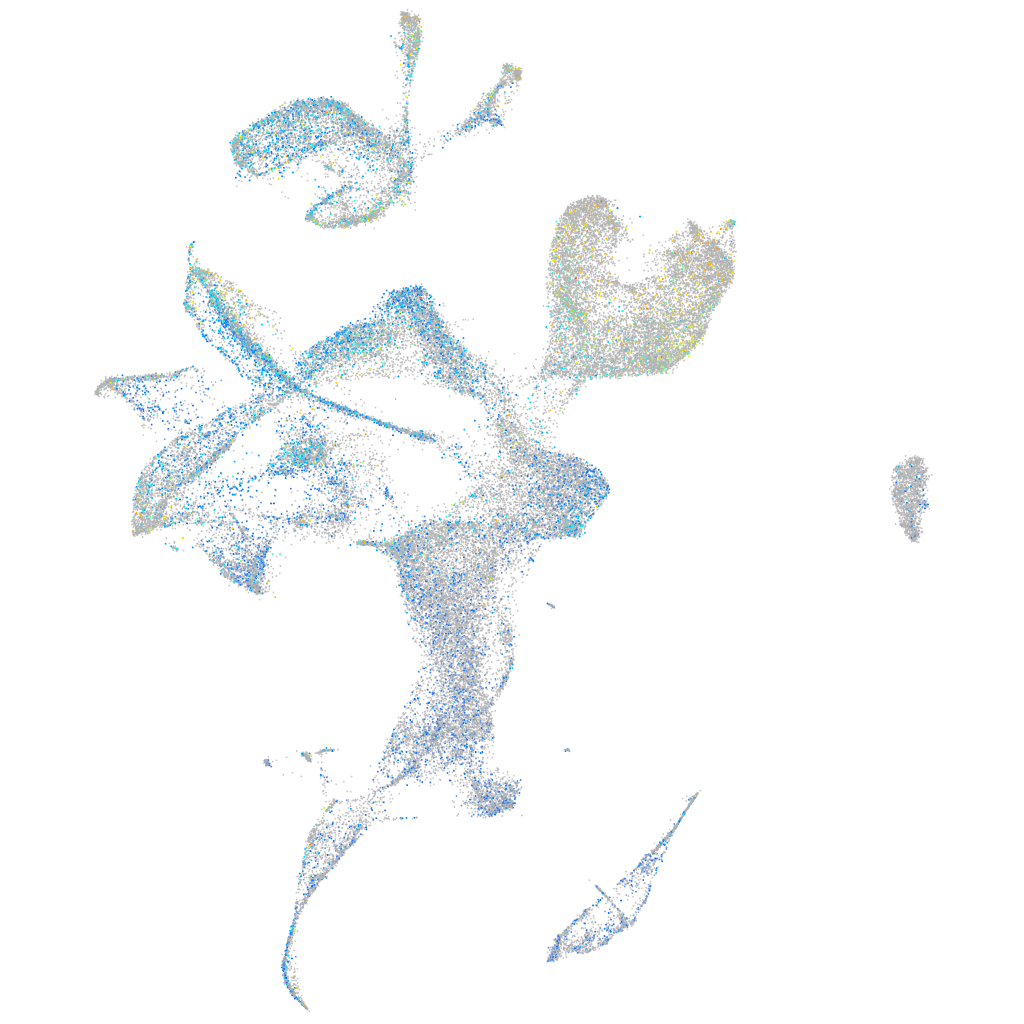
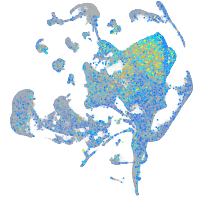
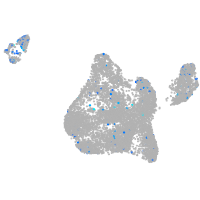
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| sncb | 0.113 | pcna | -0.093 |
| rtn1b | 0.111 | lig1 | -0.083 |
| elavl3 | 0.111 | mcm7 | -0.077 |
| gng3 | 0.110 | mcm6 | -0.076 |
| mllt11 | 0.110 | CABZ01005379.1 | -0.075 |
| stmn2a | 0.109 | chaf1a | -0.074 |
| zgc:153426 | 0.107 | rrm1 | -0.073 |
| tuba1c | 0.106 | nasp | -0.073 |
| stmn1b | 0.105 | hmgb2a | -0.073 |
| gpm6ab | 0.105 | ccnd1 | -0.072 |
| zgc:65894 | 0.105 | fen1 | -0.072 |
| rnasekb | 0.102 | banf1 | -0.072 |
| ywhag2 | 0.102 | mcm5 | -0.072 |
| ckbb | 0.101 | mcm2 | -0.072 |
| id4 | 0.101 | her15.1 | -0.072 |
| gnao1a | 0.101 | cdca7b | -0.071 |
| stxbp1a | 0.101 | mcm4 | -0.071 |
| gap43 | 0.100 | rpa3 | -0.071 |
| vamp2 | 0.100 | id1 | -0.070 |
| atp6v0cb | 0.097 | stmn1a | -0.070 |
| stx1b | 0.097 | mcm3 | -0.070 |
| ppp1r14ba | 0.096 | cdca7a | -0.069 |
| eno2 | 0.095 | rpa2 | -0.067 |
| tubb5 | 0.095 | dut | -0.067 |
| rtn1a | 0.094 | selenoh | -0.067 |
| elavl4 | 0.094 | XLOC-003690 | -0.066 |
| si:dkeyp-75h12.5 | 0.094 | esco2 | -0.065 |
| dpysl3 | 0.093 | nutf2l | -0.065 |
| islr2 | 0.092 | ranbp1 | -0.065 |
| rbpms2b | 0.092 | hells | -0.065 |
| calr | 0.092 | anp32b | -0.064 |
| stmn2b | 0.090 | slbp | -0.063 |
| carmil2 | 0.089 | rfc4 | -0.063 |
| tmsb2 | 0.089 | rrm2 | -0.061 |
| rbpms2a | 0.089 | mdka | -0.059 |