dual specificity phosphatase 23b
ZFIN 
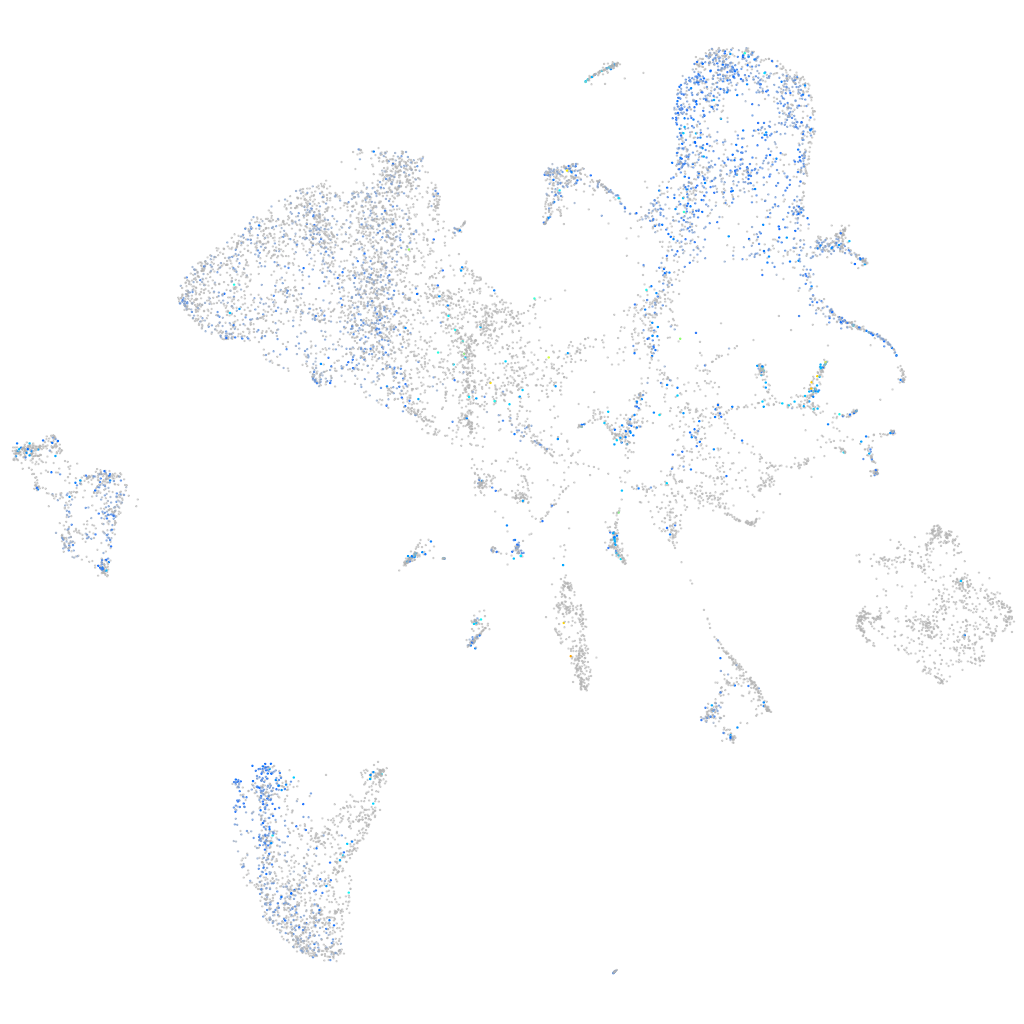
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| tm4sf4 | 0.246 | apoc1 | -0.173 |
| mgst3a | 0.243 | hspb1 | -0.132 |
| cldn15la | 0.241 | akap12b | -0.124 |
| vil1 | 0.237 | marcksb | -0.118 |
| anxa2b | 0.235 | fetub | -0.116 |
| zgc:158846 | 0.235 | cx43.4 | -0.116 |
| calml4a | 0.232 | tfa | -0.115 |
| si:dkeyp-73b11.8 | 0.230 | bhmt | -0.114 |
| zgc:172079 | 0.230 | apoa2 | -0.113 |
| zgc:77748 | 0.230 | marcksl1b | -0.112 |
| stoml3b | 0.228 | si:ch211-152c2.3 | -0.112 |
| mat2al | 0.226 | serpina1 | -0.112 |
| dhrs1 | 0.223 | zgc:123103 | -0.111 |
| plac8.1 | 0.222 | rbp4 | -0.110 |
| cdh17 | 0.220 | rbp2b | -0.109 |
| ociad2 | 0.220 | fabp10a | -0.108 |
| s100a10a | 0.219 | si:dkey-56m19.5 | -0.108 |
| si:dkey-36i7.3 | 0.219 | cdh6 | -0.108 |
| bin2a | 0.219 | serpina1l | -0.107 |
| mdh2 | 0.218 | ces2 | -0.106 |
| zgc:153968 | 0.218 | apom | -0.106 |
| GCA | 0.216 | hao1 | -0.104 |
| bpnt1 | 0.214 | fgg | -0.103 |
| zgc:136472 | 0.213 | stm | -0.103 |
| anks4b | 0.213 | ambp | -0.102 |
| serpinb1l3 | 0.213 | qkia | -0.101 |
| atp1a1a.4 | 0.212 | wu:fj16a03 | -0.101 |
| ush1c | 0.212 | cpn1 | -0.100 |
| eps8l3b | 0.212 | fgb | -0.100 |
| pdzk1 | 0.211 | ttr | -0.099 |
| aqp8a.2 | 0.211 | c9 | -0.099 |
| krt92 | 0.210 | LOC110437731 | -0.098 |
| rtn4b | 0.210 | cfhl4 | -0.097 |
| si:ch211-133l5.7 | 0.210 | gc | -0.096 |
| si:ch211-161h7.8 | 0.209 | nr6a1a | -0.096 |