dystrobrevin binding protein 1a
ZFIN 
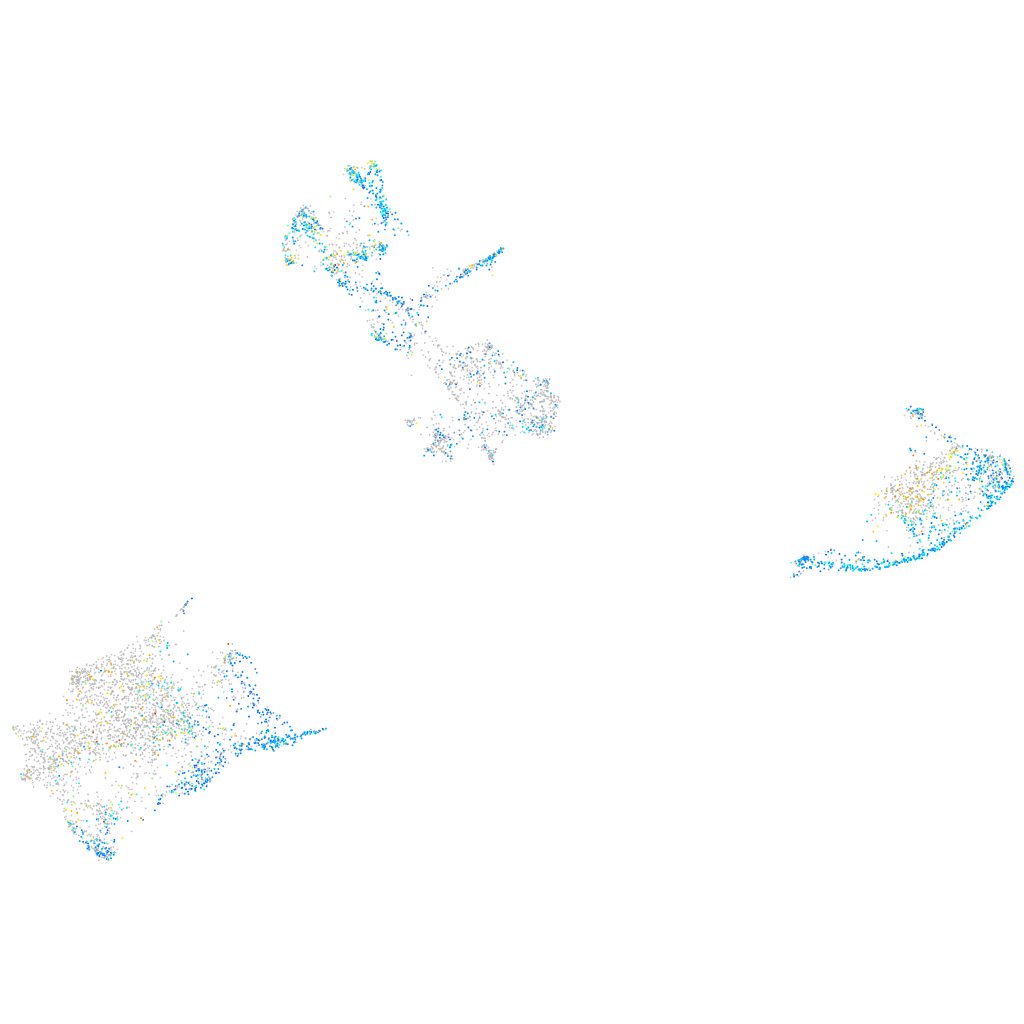
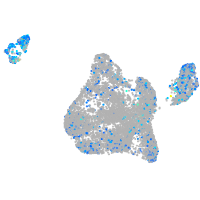
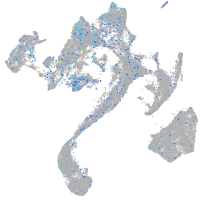
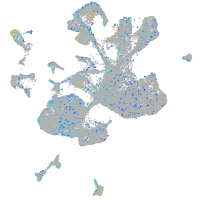
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| smim29 | 0.283 | CABZ01021592.1 | -0.248 |
| si:ch211-243a20.3 | 0.273 | si:dkey-251i10.2 | -0.213 |
| id2a | 0.270 | slc22a7a | -0.167 |
| pnp4a | 0.270 | gch2 | -0.156 |
| slc45a2 | 0.264 | hbbe1.3 | -0.142 |
| atox1 | 0.254 | aqp3a | -0.139 |
| zgc:110239 | 0.253 | hbae1.1 | -0.139 |
| LOC103909099 | 0.252 | tmem130 | -0.138 |
| sptlc2a | 0.249 | tuba1c | -0.135 |
| rab32a | 0.248 | si:ch211-251b21.1 | -0.134 |
| nme4 | 0.243 | pvalb2 | -0.133 |
| trpm1b | 0.235 | si:ch211-222l21.1 | -0.132 |
| agtrap | 0.230 | tmsb | -0.126 |
| emp3b | 0.228 | actc1b | -0.122 |
| BX571715.1 | 0.227 | hbae3 | -0.120 |
| slc37a2 | 0.227 | nova2 | -0.119 |
| CABZ01048402.2 | 0.224 | stmn1b | -0.118 |
| psph | 0.223 | ptmaa | -0.118 |
| ldhba | 0.223 | gpm6aa | -0.113 |
| ctsba | 0.220 | hbbe1.1 | -0.112 |
| zgc:110789 | 0.217 | zgc:110339 | -0.111 |
| guk1a | 0.217 | pvalb1 | -0.110 |
| atp11a | 0.215 | krt4 | -0.108 |
| anxa4 | 0.214 | fabp7a | -0.108 |
| eno3 | 0.213 | uraha | -0.106 |
| gpr143 | 0.212 | CU467822.1 | -0.106 |
| rnaseka | 0.210 | elavl3 | -0.106 |
| tfec | 0.209 | aldob | -0.104 |
| alx4a | 0.208 | marcksl1a | -0.104 |
| alx4b | 0.208 | mylpfa | -0.103 |
| si:ch73-89b15.3 | 0.207 | hmgb1b | -0.100 |
| si:ch211-256m1.8 | 0.206 | epb41a | -0.098 |
| zgc:165573 | 0.205 | cyt1 | -0.098 |
| sh3kbp1 | 0.205 | zgc:153031 | -0.098 |
| fxyd1 | 0.205 | gng3 | -0.097 |