dual serine/threonine and tyrosine protein kinase
ZFIN 
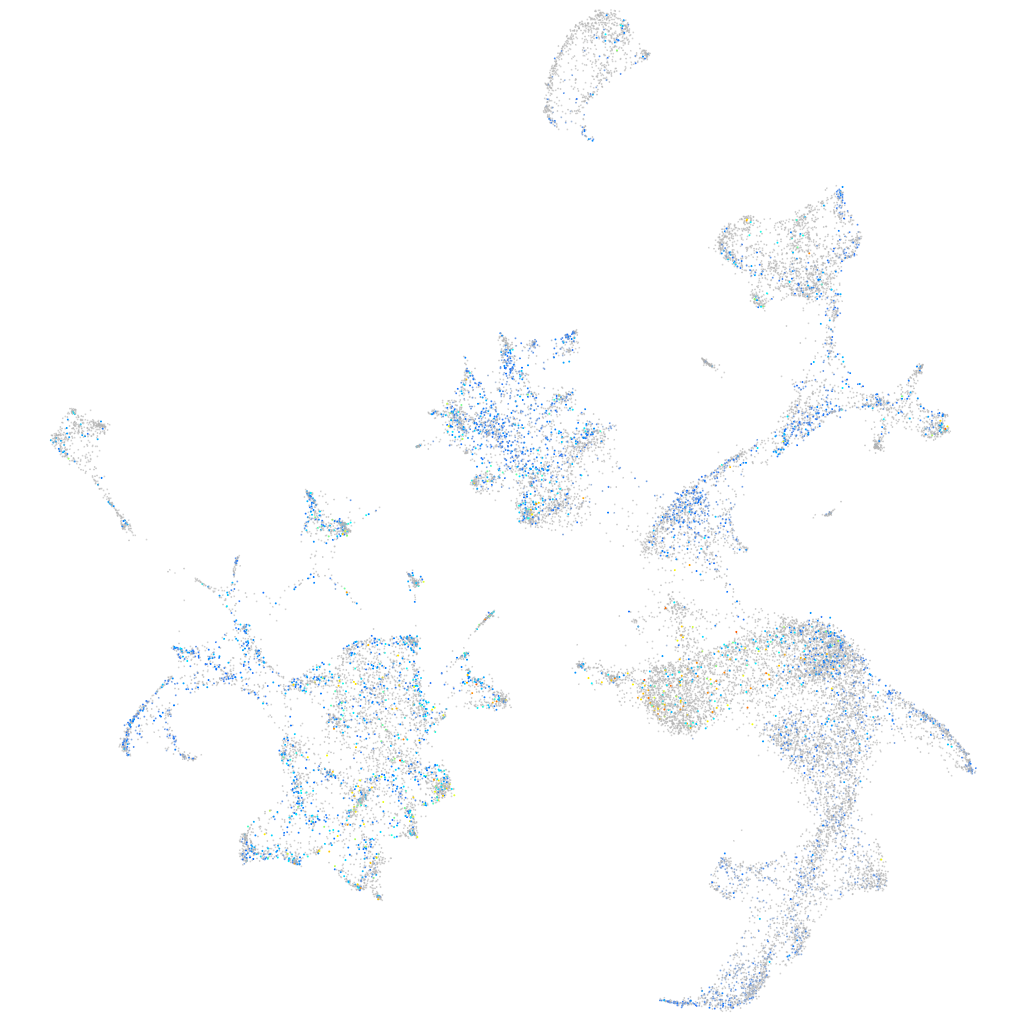
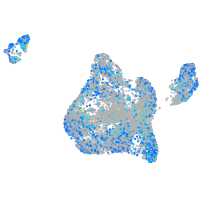
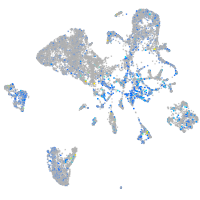
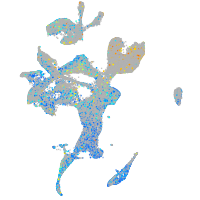
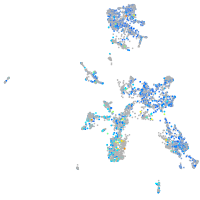
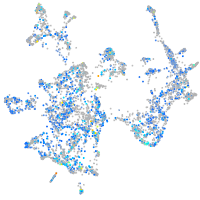
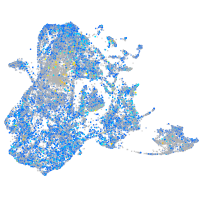
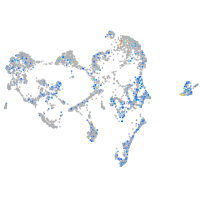
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| krt18a.1 | 0.137 | hbbe3 | -0.080 |
| krt8 | 0.137 | hbae1.3 | -0.078 |
| jpt1b | 0.135 | blvrb | -0.074 |
| tpm4a | 0.129 | hemgn | -0.074 |
| vdac3 | 0.123 | hbae1.1 | -0.073 |
| si:dkey-261h17.1 | 0.122 | zgc:163057 | -0.071 |
| qkia | 0.120 | cahz | -0.070 |
| id1 | 0.118 | hbbe1.3 | -0.070 |
| clec14a | 0.118 | hbbe1.2 | -0.068 |
| fxyd6l | 0.117 | alas2 | -0.066 |
| pmp22a | 0.117 | hbae3 | -0.065 |
| ahnak | 0.116 | hbbe1.1 | -0.065 |
| cdh5 | 0.114 | fcer1gl | -0.064 |
| pfn2l | 0.114 | slc4a1a | -0.062 |
| cavin2b | 0.113 | hbbe2 | -0.059 |
| fstl1b | 0.112 | ctss2.1 | -0.059 |
| sox7 | 0.112 | si:dkey-102g19.3 | -0.058 |
| nova2 | 0.112 | si:ch73-248e21.7 | -0.058 |
| cd81a | 0.112 | coro1a | -0.056 |
| fkbp1aa | 0.111 | tspo | -0.056 |
| akap12b | 0.111 | lect2l | -0.055 |
| egfl7 | 0.111 | prdx2 | -0.055 |
| si:ch211-156j16.1 | 0.111 | hcls1 | -0.054 |
| gpm6ab | 0.110 | laptm5 | -0.053 |
| CR383676.1 | 0.110 | tmem14ca | -0.052 |
| flot2a | 0.110 | hbae5 | -0.051 |
| cpn1 | 0.109 | mpx | -0.051 |
| dusp5 | 0.109 | alox5ap | -0.050 |
| anxa13 | 0.109 | wasb | -0.049 |
| h2afy2 | 0.108 | spi1b | -0.049 |
| gstt1b | 0.108 | lyz | -0.049 |
| col4a1 | 0.108 | npsn | -0.049 |
| her6 | 0.108 | hmbsb | -0.049 |
| marcksl1a | 0.108 | nt5c2l1 | -0.048 |
| h3f3c | 0.108 | plac8l1 | -0.048 |