desmoglein 2 like
ZFIN 
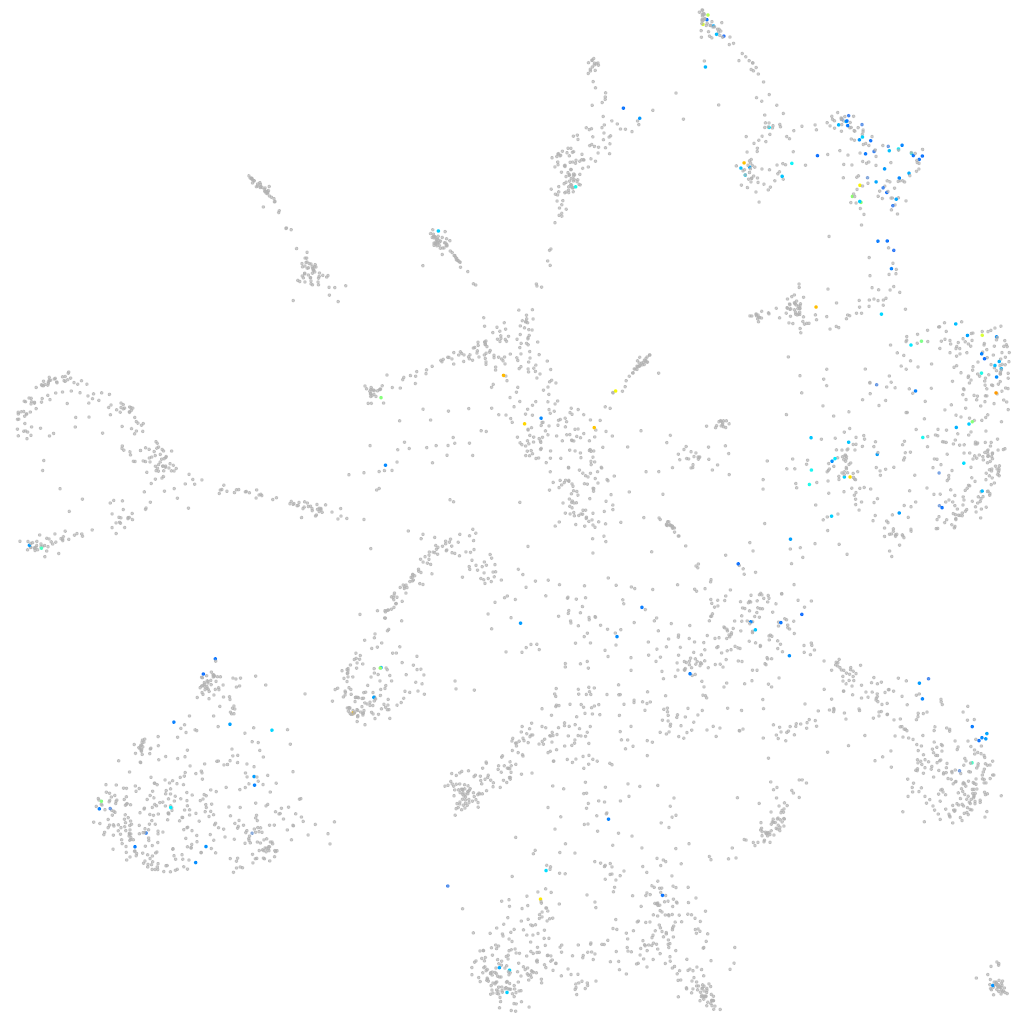
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| LOC110440062 | 0.191 | BX088707.3 | -0.105 |
| podxl | 0.179 | myl9a | -0.101 |
| fabp11a | 0.179 | ptmaa | -0.088 |
| XLOC-039672 | 0.177 | tagln | -0.084 |
| COLEC10 | 0.173 | tpm1 | -0.080 |
| XLOC-042222 | 0.172 | acta2 | -0.079 |
| tnni1b | 0.171 | zgc:153704 | -0.075 |
| sfrp5 | 0.168 | myh11a | -0.068 |
| mmel1 | 0.167 | foxf2a | -0.068 |
| tmem88b | 0.166 | mylkb | -0.067 |
| FITM1 (1 of many) | 0.166 | foxp4 | -0.067 |
| akap12b | 0.164 | gucy1b1 | -0.067 |
| jam2b | 0.163 | gapdhs | -0.065 |
| XLOC-039363 | 0.160 | ntn5 | -0.064 |
| lrp2a | 0.158 | myl6 | -0.062 |
| bnc2 | 0.157 | foxf1 | -0.060 |
| pon1 | 0.156 | csrp1b | -0.060 |
| cxadr | 0.156 | ptch2 | -0.059 |
| krt94 | 0.153 | sat1a.2 | -0.058 |
| si:dkey-157g16.6 | 0.152 | inka1a | -0.057 |
| lmod2a | 0.152 | XLOC-025423 | -0.057 |
| CR846087.3 | 0.151 | tpm2 | -0.057 |
| gata5 | 0.150 | mdkb | -0.056 |
| gata6 | 0.147 | lmod1b | -0.056 |
| myh6 | 0.145 | angptl5 | -0.056 |
| krt8 | 0.145 | LO018340.1 | -0.055 |
| epb41l5 | 0.145 | desmb | -0.055 |
| gzmk | 0.145 | lbh | -0.055 |
| tgm2b | 0.144 | inhbaa | -0.054 |
| tti1 | 0.142 | selenom | -0.054 |
| nexn | 0.141 | gpia | -0.054 |
| ftr82 | 0.141 | tdh | -0.053 |
| myh10 | 0.140 | myocd | -0.053 |
| jcada | 0.139 | cald1b | -0.053 |
| rhag | 0.138 | ndufab1b | -0.053 |