"D4, zinc and double PHD fingers family 2, like"
ZFIN 
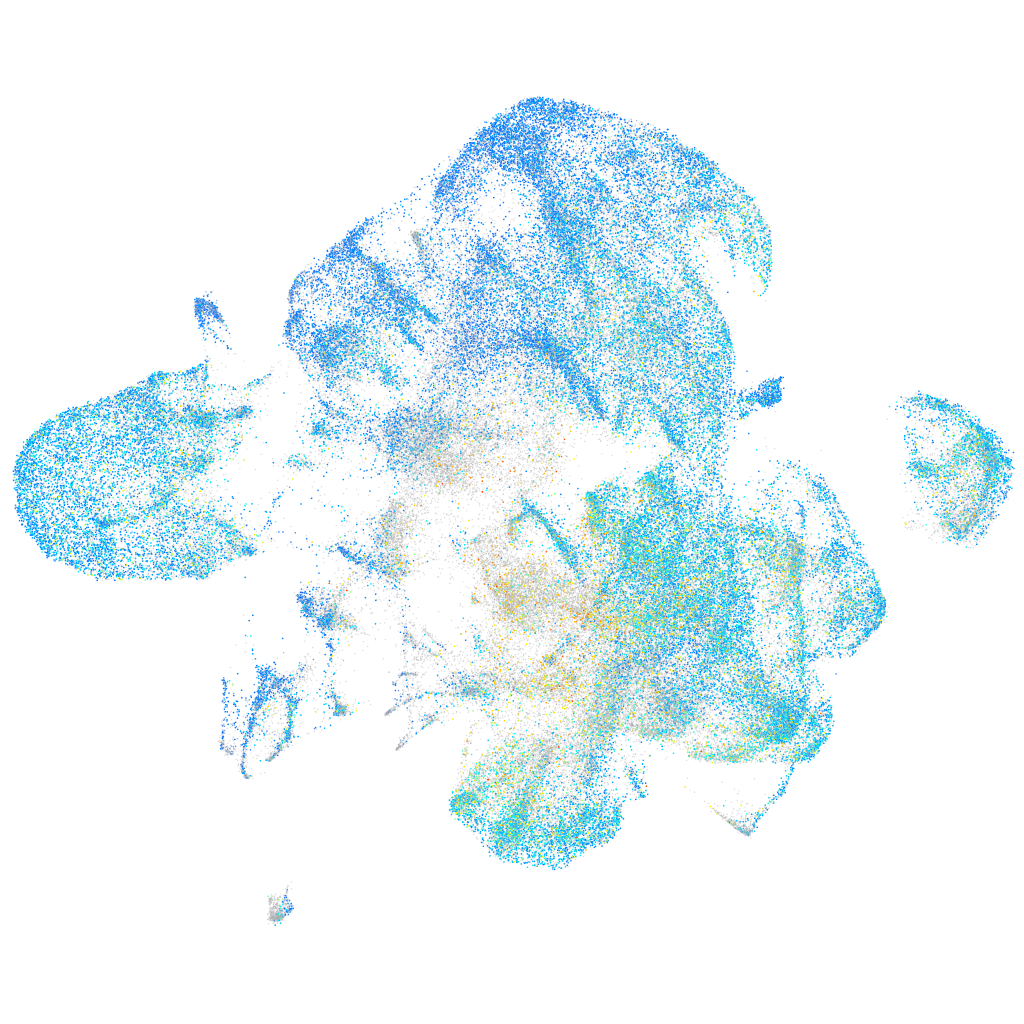
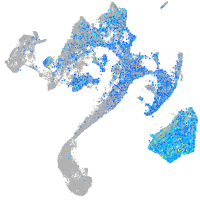
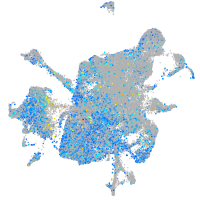
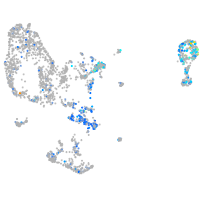
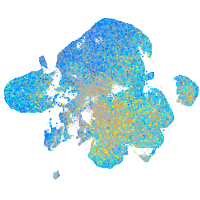
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| hnrnpaba | 0.192 | fabp7a | -0.074 |
| cirbpb | 0.180 | apoa2 | -0.066 |
| si:ch211-288g17.3 | 0.174 | atp2b2 | -0.064 |
| marcksb | 0.170 | mdka | -0.062 |
| khdrbs1a | 0.169 | dut | -0.062 |
| h3f3d | 0.167 | apoa1b | -0.060 |
| ptmab | 0.166 | zgc:56493 | -0.059 |
| hmgn6 | 0.164 | atp2b3b | -0.058 |
| hnrnpa0a | 0.163 | rpl37 | -0.058 |
| hdac1 | 0.162 | sparc | -0.057 |
| hnrnpa0b | 0.162 | nptna | -0.056 |
| smarce1 | 0.156 | eef1da | -0.056 |
| nono | 0.156 | actc1b | -0.054 |
| si:dkey-56m19.5 | 0.156 | icn | -0.054 |
| hnrnpub | 0.153 | COX7A2 (1 of many) | -0.053 |
| hnrnpabb | 0.153 | pmp22a | -0.052 |
| hp1bp3 | 0.152 | fosab | -0.052 |
| hnrnph1l | 0.150 | pabpc1b | -0.052 |
| syncrip | 0.149 | prss1 | -0.052 |
| hnrnpa1a | 0.148 | pvalb2 | -0.051 |
| si:ch211-222l21.1 | 0.148 | prss59.2 | -0.051 |
| ilf2 | 0.146 | aldocb | -0.051 |
| smarca4a | 0.145 | ctrb1 | -0.050 |
| nucks1a | 0.145 | syngr1a | -0.050 |
| chd4a | 0.143 | slc6a17 | -0.049 |
| tardbp | 0.142 | pvalb1 | -0.049 |
| ctnnbip1 | 0.141 | sv2a | -0.049 |
| rbm4.3 | 0.140 | zgc:158463 | -0.049 |
| ilf3b | 0.140 | atp1a1b | -0.048 |
| hmgb3a | 0.139 | calm1b | -0.048 |
| sumo3a | 0.138 | mt-atp8 | -0.048 |
| marcksl1b | 0.137 | egr1 | -0.047 |
| hmgb1b | 0.135 | si:ch211-156b7.4 | -0.047 |
| ssbp3b | 0.134 | sypa | -0.047 |
| pnrc2 | 0.133 | chchd10 | -0.047 |