dedicator of cytokinesis 11
ZFIN 
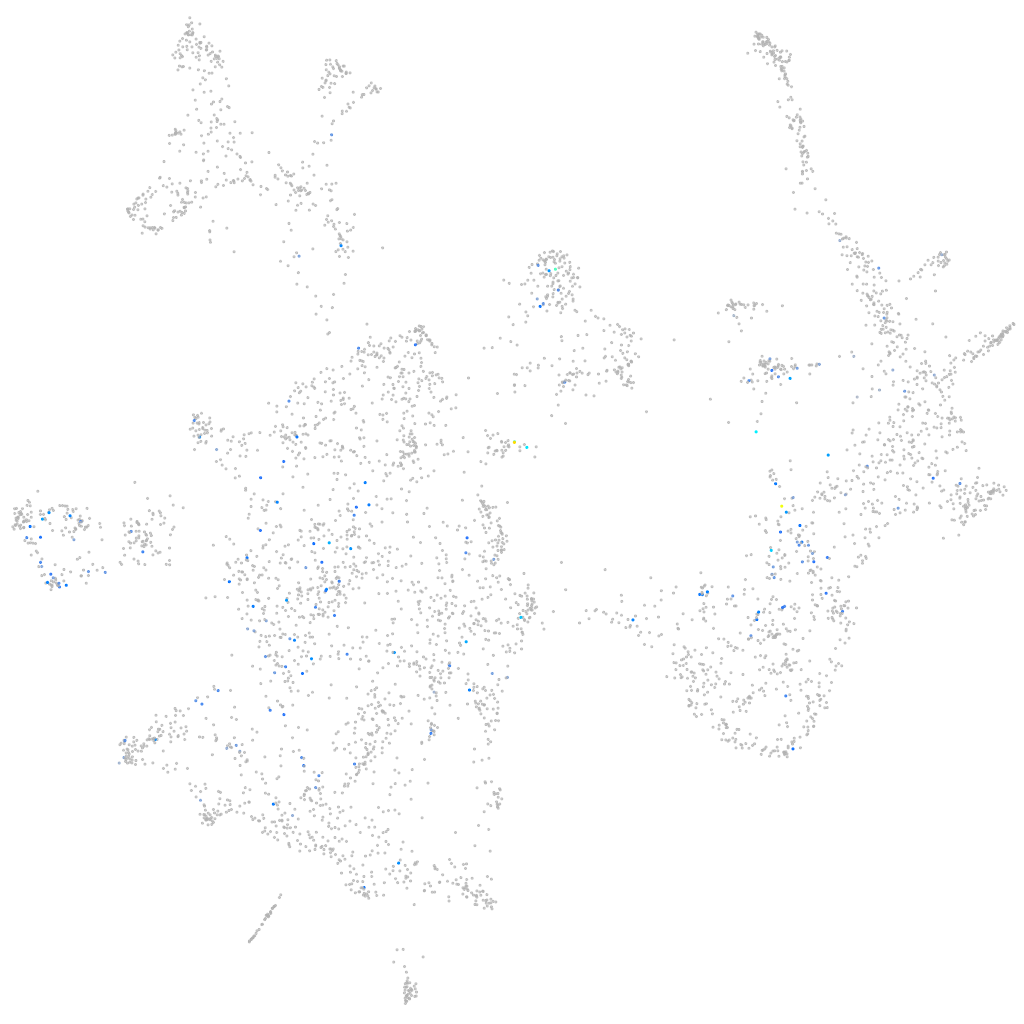
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| si:rp71-39b20.4 | 0.226 | h1f0 | -0.088 |
| camk1ga | 0.192 | pkig | -0.073 |
| FP085398.1 | 0.176 | tob1b | -0.070 |
| sult5a1 | 0.176 | s100t | -0.067 |
| XLOC-016099 | 0.175 | otofb | -0.066 |
| si:ch211-241b2.5 | 0.174 | tmem59 | -0.065 |
| tek | 0.171 | calml4a | -0.064 |
| LOC100001846 | 0.168 | pvalb8 | -0.064 |
| XLOC-043043 | 0.164 | itm2ba | -0.063 |
| XLOC-032001 | 0.163 | ap3s1.1 | -0.063 |
| CABZ01064771.1 | 0.161 | calm1a | -0.063 |
| hist1h2a4 | 0.160 | cd164l2 | -0.063 |
| XLOC-032577 | 0.158 | gpx2 | -0.063 |
| si:dkeyp-97a10.2 | 0.154 | aldocb | -0.062 |
| LOC108182890 | 0.151 | pcsk5a | -0.062 |
| kcnj1b | 0.149 | vamp8 | -0.062 |
| ccr10 | 0.149 | capgb | -0.062 |
| cygb2 | 0.149 | nptna | -0.061 |
| thap12b | 0.145 | enosf1 | -0.061 |
| zgc:158659 | 0.144 | CR925719.1 | -0.061 |
| XLOC-042505 | 0.142 | prr15la | -0.060 |
| si:dkey-7i4.8 | 0.142 | gapdhs | -0.060 |
| dut | 0.141 | hsd17b7 | -0.060 |
| LOC553308 | 0.141 | atp2b1a | -0.060 |
| stmn1a | 0.140 | atp1a3b | -0.060 |
| LO018168.1 | 0.140 | id2a | -0.059 |
| si:ch211-246b8.5 | 0.140 | gipc3 | -0.059 |
| sst2 | 0.138 | eno1a | -0.059 |
| vav3a | 0.138 | osbpl1a | -0.058 |
| chaf1a | 0.137 | enkur | -0.058 |
| cox6a2 | 0.137 | dnajc5b | -0.057 |
| znf1052 | 0.137 | emb | -0.057 |
| plxnc1 | 0.136 | tbc1d14 | -0.057 |
| ephx4 | 0.136 | gb:eh456644 | -0.056 |
| pcna | 0.136 | lrrc73 | -0.056 |