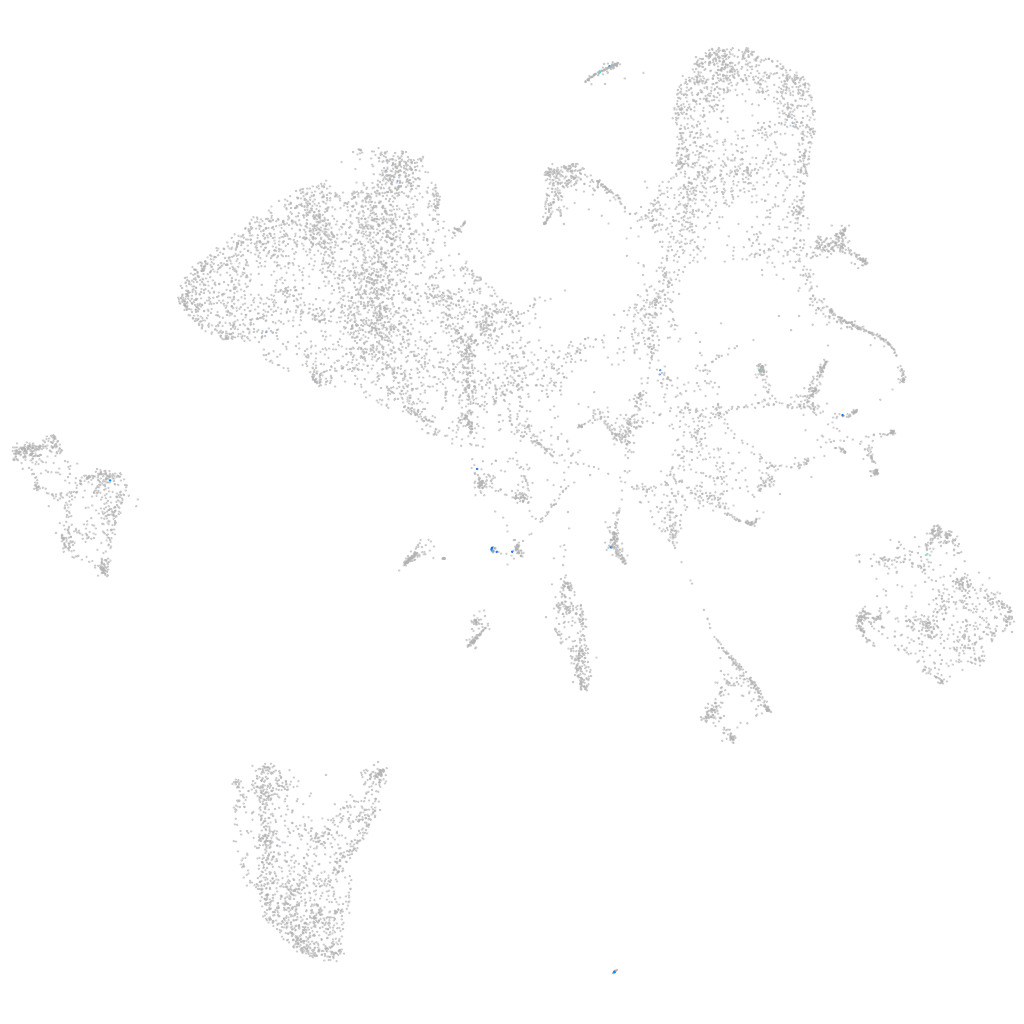
"dynein, axonemal, heavy chain 12"
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm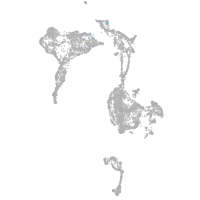 |
muscle 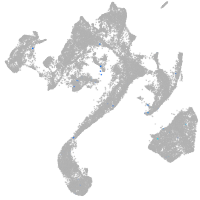 |
mural cells / non-skeletal muscle 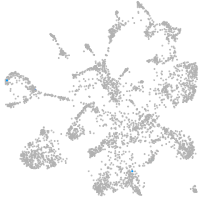 |
mesenchyme 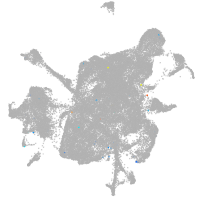 |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| CT009714.1 | 0.400 | ahcy | -0.038 |
| si:ch211-270n8.4 | 0.301 | nupr1b | -0.037 |
| chrnb3a | 0.224 | gatm | -0.036 |
| mycbpap | 0.222 | gamt | -0.035 |
| c7a | 0.210 | gapdh | -0.033 |
| LOC101886593 | 0.207 | dap | -0.032 |
| tsga10 | 0.207 | fabp3 | -0.031 |
| cfap161 | 0.206 | apoc2 | -0.031 |
| XLOC-042788 | 0.204 | bhmt | -0.031 |
| b3gat1b | 0.204 | aqp12 | -0.030 |
| spata18 | 0.199 | krtcap2 | -0.030 |
| si:dkey-151j17.4 | 0.199 | afp4 | -0.030 |
| si:dkey-182i3.8 | 0.198 | fbp1b | -0.029 |
| armc4 | 0.198 | sec61a1 | -0.029 |
| si:ch73-44m9.2 | 0.196 | adka | -0.028 |
| LOC103911624 | 0.195 | mat1a | -0.028 |
| wisp1b | 0.191 | gstt1a | -0.028 |
| gmnc | 0.190 | aldh7a1 | -0.027 |
| syt17 | 0.189 | acmsd | -0.027 |
| stmnd1 | 0.184 | cx32.3 | -0.027 |
| zgc:195356 | 0.184 | hspe1 | -0.027 |
| ctxn2 | 0.178 | apoa4b.1 | -0.026 |
| oprm1 | 0.176 | slc16a10 | -0.026 |
| cfap57 | 0.175 | mt-nd3 | -0.026 |
| tbata | 0.169 | cox6b1 | -0.026 |
| LOC108182168 | 0.168 | uqcrfs1 | -0.026 |
| LOC101886954 | 0.168 | ssr3 | -0.026 |
| LOC101885342 | 0.168 | pck2 | -0.026 |
| si:ch211-130h14.4 | 0.166 | gpx1a | -0.026 |
| LOC108179803 | 0.165 | abat | -0.026 |
| CR749763.7 | 0.164 | nipsnap3a | -0.026 |
| ropn1l | 0.163 | rdh1 | -0.025 |
| slc8a1b | 0.163 | wu:fj16a03 | -0.025 |
| cfap52 | 0.163 | mdh1aa | -0.025 |
| fhdc1 | 0.163 | aldh6a1 | -0.025 |