delta-like 4 (Drosophila)
ZFIN 
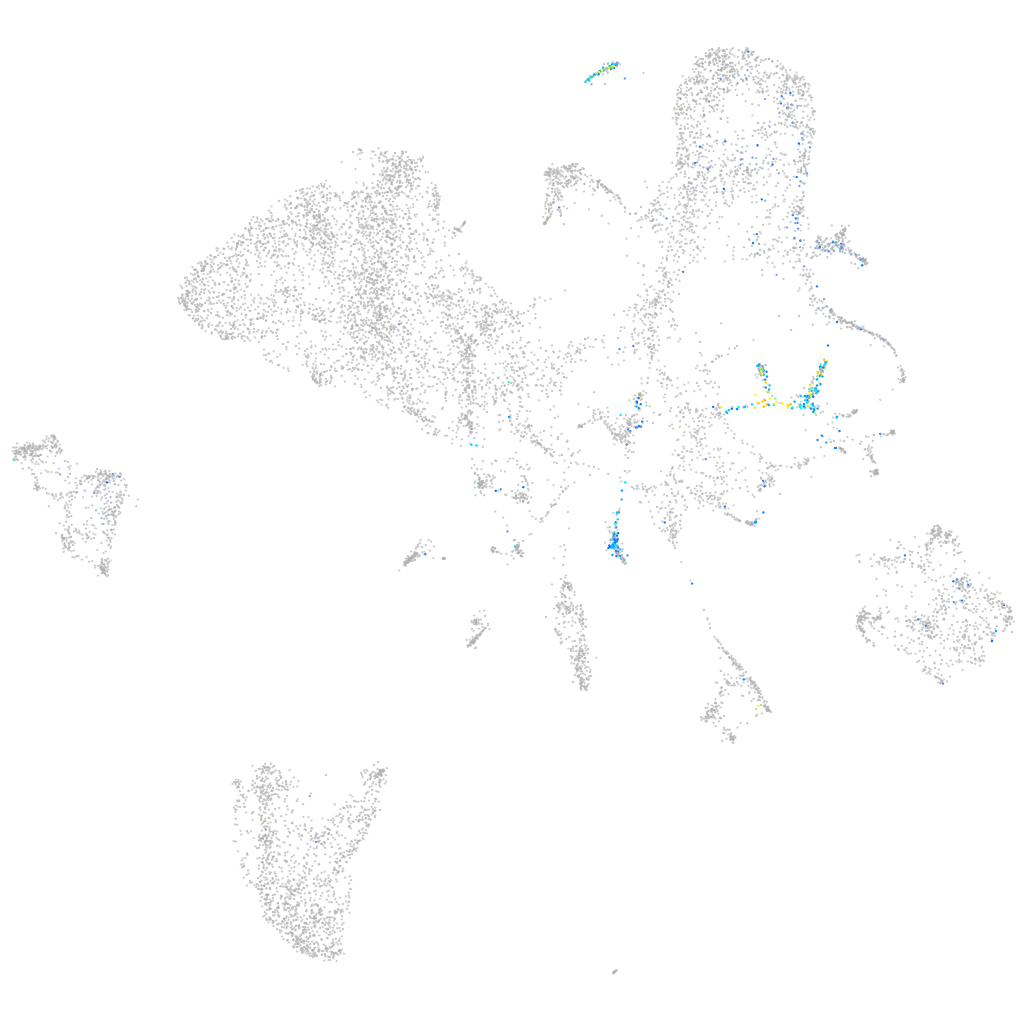
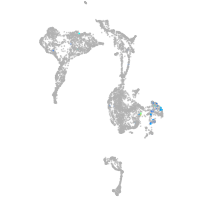
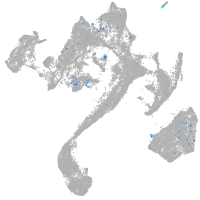
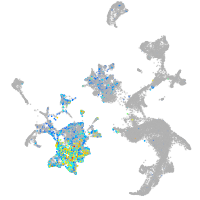
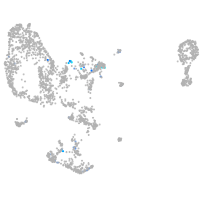
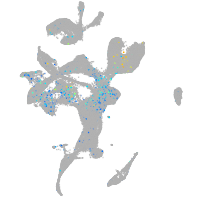
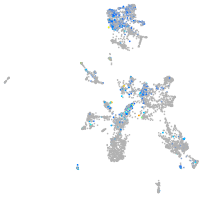
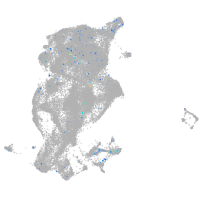
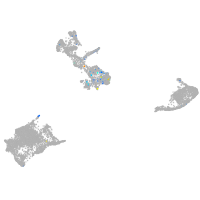
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| insm1b | 0.503 | fabp3 | -0.175 |
| scg3 | 0.494 | gamt | -0.170 |
| neurod1 | 0.466 | ahcy | -0.166 |
| fev | 0.457 | bhmt | -0.152 |
| egr4 | 0.448 | gatm | -0.148 |
| hepacam2 | 0.425 | mat1a | -0.145 |
| zgc:101731 | 0.396 | gapdh | -0.145 |
| mir375-2 | 0.386 | hdlbpa | -0.140 |
| mir7a-1 | 0.386 | aqp12 | -0.139 |
| arxa | 0.367 | apoa1b | -0.137 |
| c2cd4a | 0.362 | apoc1 | -0.132 |
| anxa4 | 0.358 | nupr1b | -0.131 |
| ret | 0.355 | agxtb | -0.131 |
| capsla | 0.348 | adka | -0.130 |
| id4 | 0.348 | aldh7a1 | -0.130 |
| pax4 | 0.348 | aldh6a1 | -0.130 |
| pcsk1nl | 0.343 | fbp1b | -0.128 |
| nkx2.2a | 0.341 | si:dkey-16p21.8 | -0.127 |
| elovl1a | 0.326 | tfa | -0.127 |
| slc7a14a | 0.323 | cx32.3 | -0.127 |
| lmx1ba | 0.319 | apoa2 | -0.125 |
| tox | 0.318 | afp4 | -0.125 |
| si:ch73-160i9.2 | 0.317 | apoc2 | -0.124 |
| adcyap1a | 0.315 | apoa4b.1 | -0.124 |
| insl5a | 0.312 | wu:fj16a03 | -0.123 |
| vat1 | 0.310 | agxta | -0.123 |
| calm1b | 0.309 | mgst1.2 | -0.120 |
| pax6b | 0.307 | serpina1 | -0.119 |
| tmsb | 0.304 | gstt1a | -0.119 |
| zgc:165481 | 0.304 | ttc36 | -0.119 |
| rasd4 | 0.304 | serpina1l | -0.118 |
| adgrg4b | 0.302 | ftcd | -0.117 |
| penka | 0.301 | rbp2b | -0.117 |
| nmu | 0.297 | apom | -0.117 |
| lysmd2 | 0.295 | serpinf2b | -0.117 |