"discs, large (Drosophila) homolog-associated protein 4a"
ZFIN 
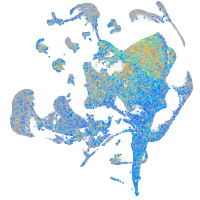
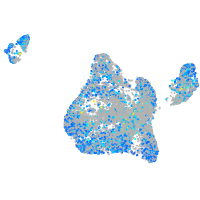
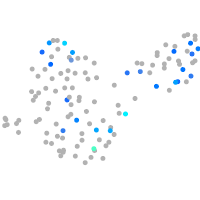
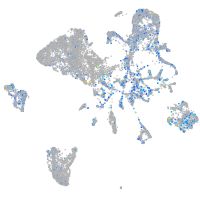
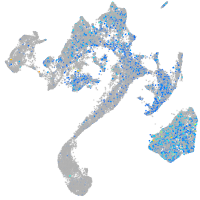
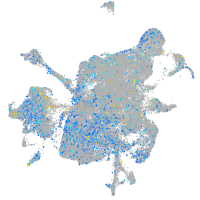
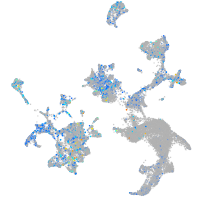
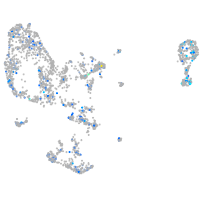
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| zgc:110540 | 0.157 | atp1b1a | -0.045 |
| CABZ01005379.1 | 0.139 | elavl3 | -0.045 |
| slbp | 0.136 | h2afx1 | -0.044 |
| chaf1a | 0.135 | fabp11a | -0.042 |
| pcna | 0.134 | adgrl3.1 | -0.042 |
| tk1 | 0.133 | ppdpfb | -0.042 |
| rrm2 | 0.133 | dct | -0.040 |
| mibp | 0.131 | pmela | -0.040 |
| dut | 0.126 | slc45a2 | -0.040 |
| nasp | 0.126 | rbpms2b | -0.039 |
| esco2 | 0.123 | slc3a2a | -0.039 |
| fen1 | 0.122 | rbfox2 | -0.039 |
| rrm1 | 0.122 | igfbp7 | -0.039 |
| asf1bb | 0.121 | tyrp1b | -0.039 |
| stmn1a | 0.121 | col4a5 | -0.038 |
| mcm7 | 0.121 | rbp3 | -0.038 |
| atad2 | 0.118 | sncb | -0.038 |
| rpa3 | 0.117 | crybb1 | -0.037 |
| rfc3 | 0.116 | scrt2 | -0.037 |
| dek | 0.116 | barhl2 | -0.037 |
| rpa2 | 0.115 | zgc:73359 | -0.037 |
| asf1ba | 0.115 | pttg1ipb | -0.037 |
| btg3 | 0.114 | mir181b-3 | -0.036 |
| hmgb2a | 0.114 | gngt2b | -0.036 |
| XLOC-042899 | 0.114 | cryba2a | -0.036 |
| dck | 0.111 | cryba1b | -0.036 |
| e2f7 | 0.111 | tspan36 | -0.036 |
| ccne2 | 0.109 | cryba4 | -0.036 |
| baz1b | 0.109 | sparc | -0.036 |
| dnajc9 | 0.106 | gnat2 | -0.035 |
| prim2 | 0.106 | pmelb | -0.035 |
| lig1 | 0.106 | tyrp1a | -0.035 |
| rfc4 | 0.106 | rbp4l | -0.035 |
| suv39h1b | 0.105 | tinagl1 | -0.035 |
| seta | 0.105 | tcima | -0.035 |