dihydrolipoamide dehydrogenase
ZFIN 
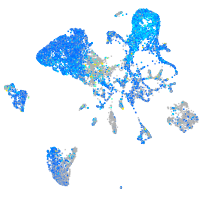
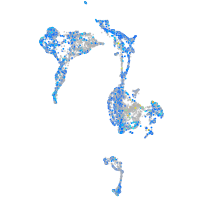
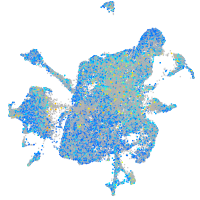
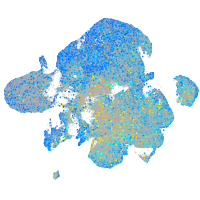
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| atp5if1b | 0.465 | si:ch73-1a9.3 | -0.399 |
| mdh2 | 0.455 | ptmab | -0.389 |
| atp5f1b | 0.451 | pabpc1a | -0.376 |
| aldoab | 0.448 | si:ch211-222l21.1 | -0.372 |
| atp5meb | 0.445 | hsp90ab1 | -0.363 |
| cox7a2a | 0.429 | hmgb2b | -0.351 |
| gapdh | 0.429 | hmgb2a | -0.347 |
| eno3 | 0.429 | h3f3d | -0.346 |
| atp5mc3b | 0.426 | cirbpa | -0.342 |
| atp5fa1 | 0.424 | si:ch73-281n10.2 | -0.340 |
| atp5mc1 | 0.420 | hnrnpaba | -0.336 |
| nme2b.2 | 0.417 | hmga1a | -0.329 |
| pvalb4 | 0.417 | khdrbs1a | -0.328 |
| got2b | 0.417 | anp32a | -0.326 |
| suclg1 | 0.416 | h2afvb | -0.324 |
| got1 | 0.415 | nucks1a | -0.321 |
| atp2a1 | 0.415 | hnrnpa0b | -0.318 |
| pgam2 | 0.415 | cirbpb | -0.317 |
| aco2 | 0.413 | anp32b | -0.316 |
| sucla2 | 0.413 | hmgn7 | -0.315 |
| ttn.2 | 0.413 | seta | -0.312 |
| chchd10 | 0.411 | hnrnpabb | -0.311 |
| srl | 0.409 | fthl27 | -0.310 |
| ttn.1 | 0.408 | hmgn2 | -0.308 |
| casq2 | 0.408 | tuba8l4 | -0.308 |
| ckmb | 0.408 | hmgn6 | -0.307 |
| ckmt2b | 0.407 | cx43.4 | -0.306 |
| tmem38a | 0.407 | si:ch211-288g17.3 | -0.304 |
| atp1a2a | 0.406 | syncrip | -0.302 |
| pgk1 | 0.404 | acin1a | -0.300 |
| cox5aa | 0.403 | marcksb | -0.298 |
| ugp2a | 0.403 | cbx3a | -0.296 |
| ckma | 0.403 | hnrnpa1b | -0.294 |
| nnt | 0.403 | hnrnpa1a | -0.280 |
| actc1b | 0.402 | marcksl1b | -0.278 |