"dicer 1, ribonuclease type III"
ZFIN 
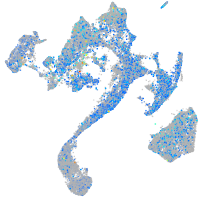
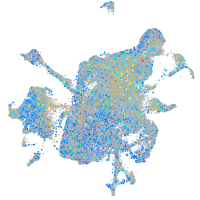
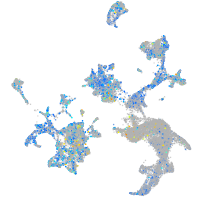
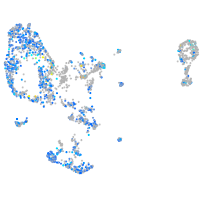
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| gsc | 0.153 | si:dkey-151g10.6 | -0.064 |
| chd4a | 0.072 | rpl39 | -0.047 |
| fgfr2 | 0.072 | rps29 | -0.047 |
| COX3 | 0.069 | rpl37 | -0.044 |
| atrx | 0.069 | rplp1 | -0.041 |
| mt-co2 | 0.067 | rpl29 | -0.039 |
| mt-cyb | 0.067 | rpl36a | -0.038 |
| mt-atp6 | 0.067 | rpl38 | -0.038 |
| rarga | 0.065 | mia | -0.036 |
| tox | 0.065 | rpl26 | -0.034 |
| mt-nd1 | 0.064 | snorc | -0.033 |
| ctnnb1 | 0.063 | rps15a | -0.031 |
| kdm6bb | 0.063 | pcolceb | -0.031 |
| celf2 | 0.063 | rps23 | -0.031 |
| mt-nd2 | 0.063 | rpl7 | -0.029 |
| tenm3 | 0.062 | rps24 | -0.028 |
| foxp4 | 0.062 | rps28 | -0.027 |
| h3f3c | 0.061 | vwde | -0.027 |
| gatad2b | 0.060 | tnn | -0.027 |
| mt-nd4 | 0.060 | rps20 | -0.027 |
| ppiaa | 0.059 | tuba8l2 | -0.027 |
| pleca | 0.059 | ptgdsa | -0.027 |
| sfrp2 | 0.058 | pvalb1 | -0.026 |
| hnrnpa0a | 0.057 | thbs1b | -0.026 |
| hp1bp3 | 0.057 | cthrc1a | -0.026 |
| meis1b | 0.056 | prss59.2 | -0.026 |
| NC-002333.4 | 0.056 | rps21 | -0.026 |
| efnb2b | 0.056 | epyc | -0.026 |
| CU929237.1 | 0.055 | acana | -0.025 |
| mt-co1 | 0.055 | rpl23 | -0.025 |
| ammecr1 | 0.055 | actc1b | -0.025 |
| meis2b | 0.055 | prss1 | -0.025 |
| fam168a | 0.055 | rpl27a | -0.024 |
| acin1a | 0.055 | VIT | -0.024 |
| efnb3b | 0.054 | aldob | -0.024 |