DET1 partner of COP1 E3 ubiquitin ligase
ZFIN 
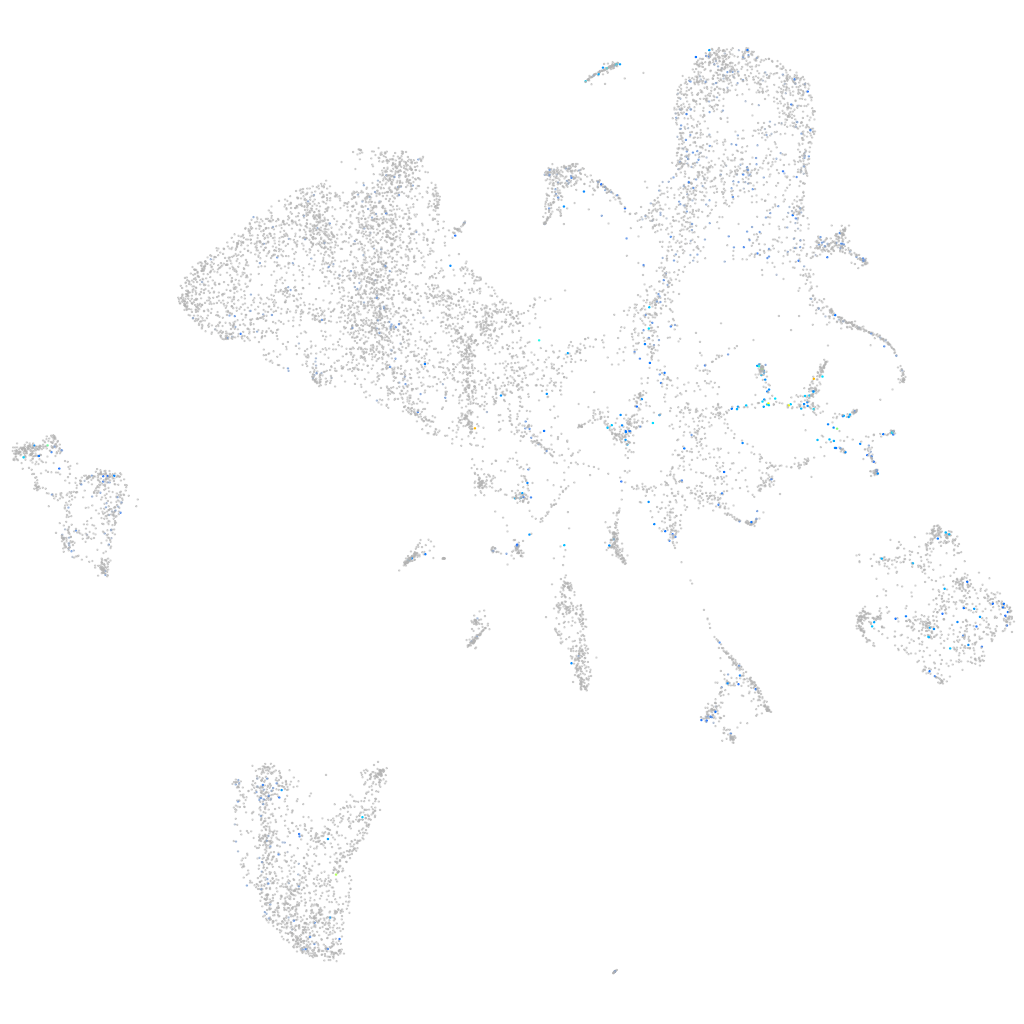
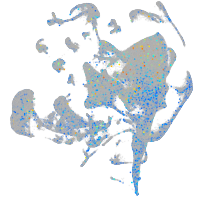
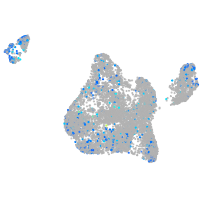
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| pygma | 0.209 | gamt | -0.112 |
| mir7a-1 | 0.203 | bhmt | -0.110 |
| scg3 | 0.199 | apoa2 | -0.103 |
| neurod1 | 0.197 | gapdh | -0.102 |
| pax6b | 0.193 | apoa1b | -0.101 |
| pax4 | 0.187 | gatm | -0.099 |
| fev | 0.185 | agxtb | -0.099 |
| mir375-2 | 0.184 | tfa | -0.097 |
| neurog3 | 0.183 | mat1a | -0.095 |
| insm1b | 0.162 | rbp2b | -0.095 |
| insm1a | 0.161 | fabp10a | -0.095 |
| tradv31.0 | 0.160 | serpina1 | -0.094 |
| nkx2.2a | 0.155 | apoc1 | -0.094 |
| ret | 0.155 | rbp4 | -0.093 |
| lysmd2 | 0.154 | apom | -0.093 |
| runx1t1 | 0.149 | serpina1l | -0.092 |
| id4 | 0.147 | ces2 | -0.092 |
| rprmb | 0.146 | fetub | -0.092 |
| dll4 | 0.145 | zgc:123103 | -0.091 |
| ddx5 | 0.143 | LOC110437731 | -0.091 |
| hepacam2 | 0.142 | uox | -0.091 |
| cacna1c | 0.142 | wu:fj16a03 | -0.090 |
| si:dkey-42i9.4 | 0.142 | hpda | -0.090 |
| nucb2b | 0.141 | pnp4b | -0.090 |
| hpcal1 | 0.141 | apoa4b.1 | -0.090 |
| elovl1a | 0.140 | ttr | -0.090 |
| si:dkey-153k10.9 | 0.139 | ambp | -0.090 |
| gfra3 | 0.138 | fgb | -0.089 |
| gnao1a | 0.137 | fgg | -0.089 |
| vat1 | 0.136 | hao1 | -0.088 |
| zgc:101731 | 0.136 | gc | -0.088 |
| tmtops3b | 0.133 | afp4 | -0.087 |
| WDR63 | 0.133 | ahcy | -0.087 |
| c2cd4a | 0.133 | c9 | -0.086 |
| CU571064.1 | 0.132 | f2 | -0.085 |