desumoylating isopeptidase 1a
ZFIN 
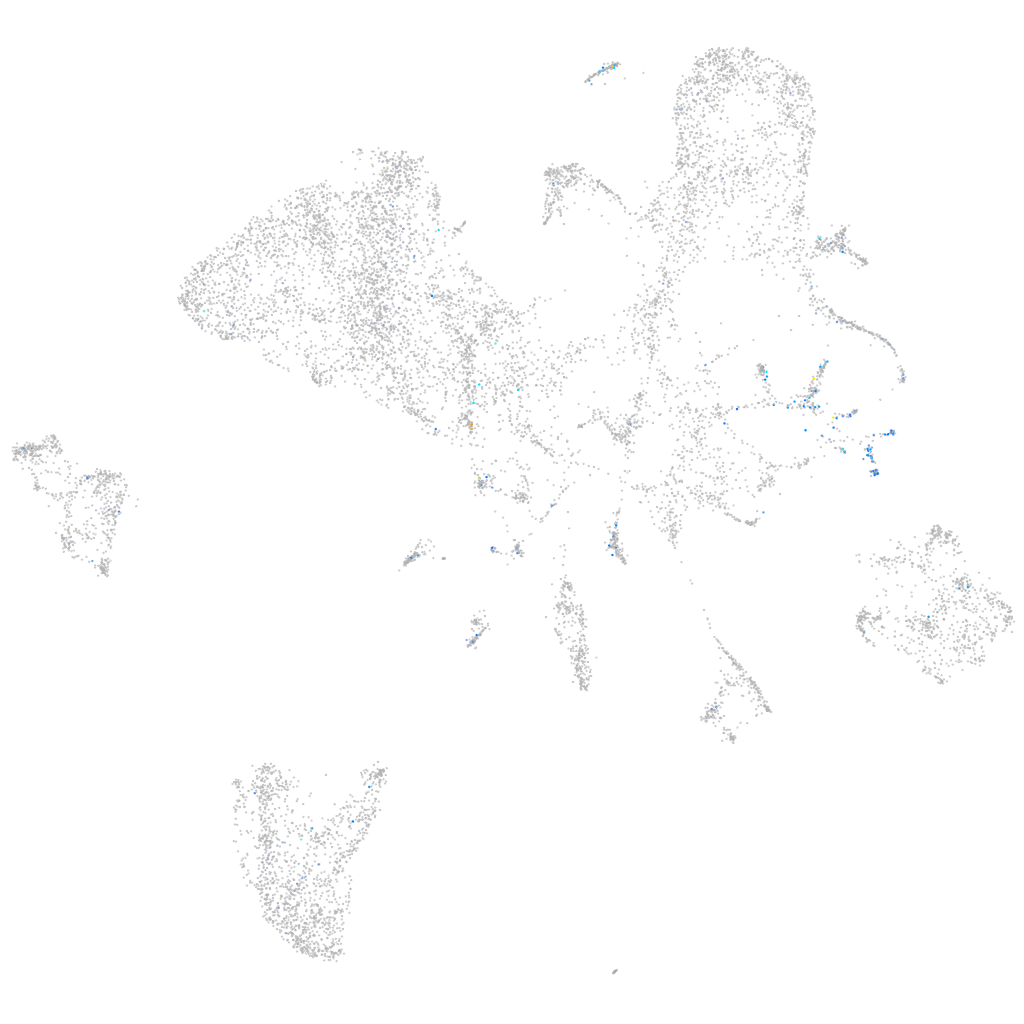
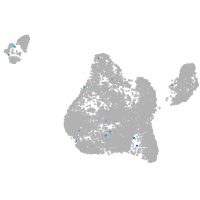
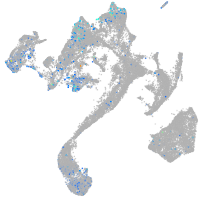
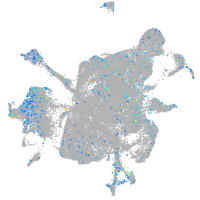
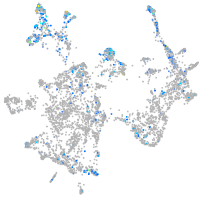
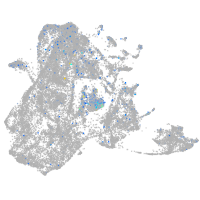
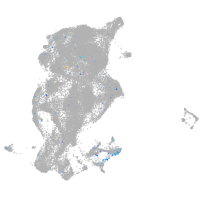
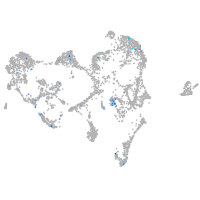
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| scg3 | 0.264 | ahcy | -0.087 |
| scgn | 0.239 | gamt | -0.086 |
| neurod1 | 0.236 | gapdh | -0.084 |
| rprmb | 0.235 | fbp1b | -0.080 |
| pcsk1 | 0.232 | gatm | -0.076 |
| syt11a | 0.223 | gstr | -0.074 |
| rims2a | 0.221 | aldob | -0.074 |
| pcsk1nl | 0.221 | apoa4b.1 | -0.073 |
| tspan7b | 0.215 | apoc2 | -0.073 |
| si:dkey-153k10.9 | 0.214 | hdlbpa | -0.072 |
| XLOC-029934 | 0.211 | adka | -0.071 |
| scg2a | 0.210 | rps20 | -0.070 |
| gpc1a | 0.210 | dap | -0.069 |
| pax6b | 0.207 | gstt1a | -0.068 |
| kcnj11 | 0.207 | aldh7a1 | -0.067 |
| cpe | 0.206 | suclg2 | -0.066 |
| syt1a | 0.205 | aldh9a1a.1 | -0.065 |
| LO018020.2 | 0.205 | apobb.1 | -0.065 |
| aldocb | 0.203 | mat1a | -0.065 |
| mir7a-1 | 0.199 | cx32.3 | -0.064 |
| vamp2 | 0.197 | apoa1b | -0.064 |
| insm1a | 0.196 | gcshb | -0.064 |
| ptchd4 | 0.196 | bhmt | -0.064 |
| cnrip1a | 0.192 | si:dkey-16p21.8 | -0.064 |
| lysmd2 | 0.191 | scp2a | -0.063 |
| pcsk2 | 0.191 | afp4 | -0.063 |
| LOC110437922 | 0.190 | hspd1 | -0.062 |
| egr4 | 0.190 | apoc1 | -0.061 |
| rnasekb | 0.189 | mgst1.2 | -0.061 |
| kcnk9 | 0.189 | msrb2 | -0.060 |
| cacng4b | 0.189 | nipsnap3a | -0.060 |
| atp6v0cb | 0.189 | CABZ01032488.1 | -0.060 |
| isl1 | 0.188 | rgn | -0.060 |
| zgc:101731 | 0.185 | aqp12 | -0.059 |
| olfm2a | 0.185 | fabp2 | -0.059 |