dCMP deaminase
ZFIN 
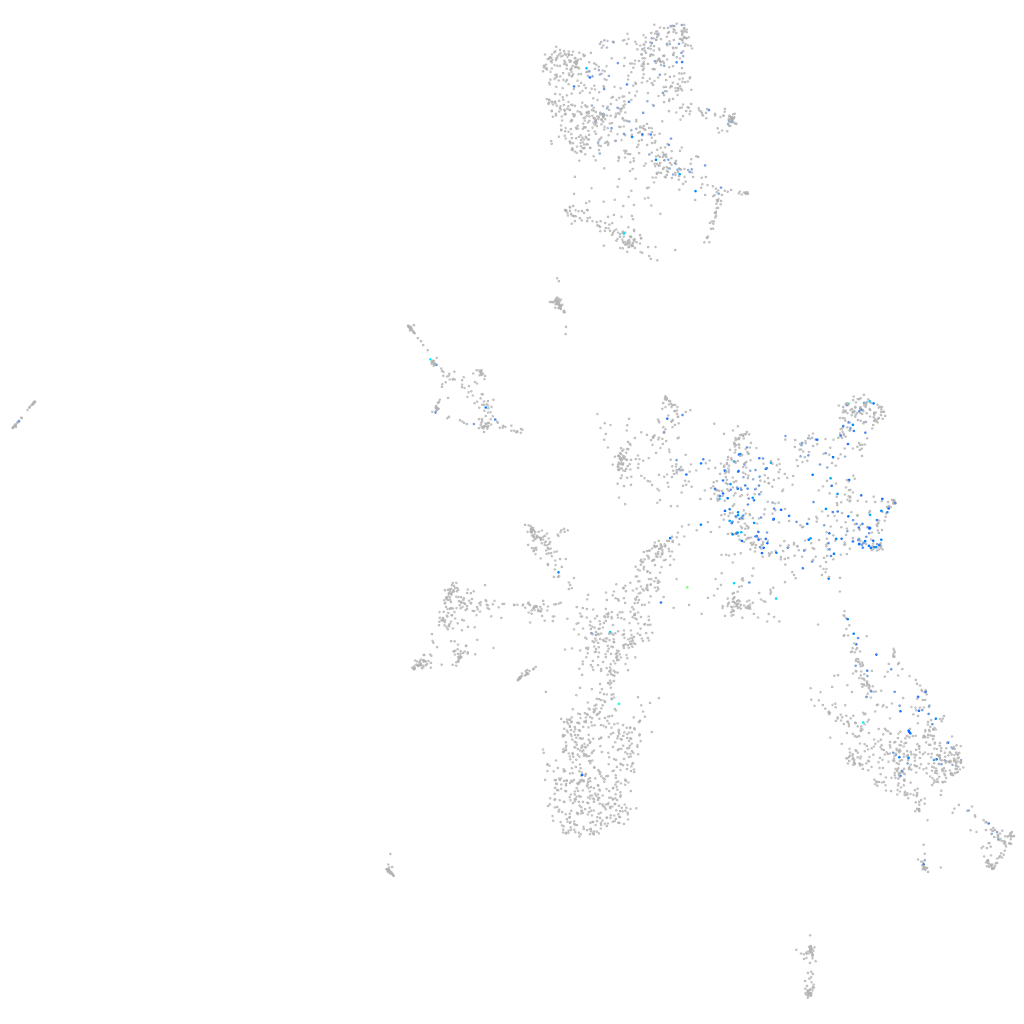
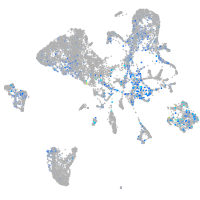
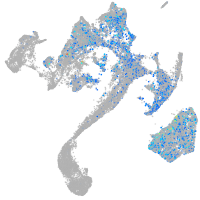
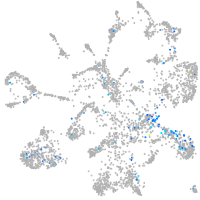
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| dut | 0.340 | txn | -0.182 |
| pcna | 0.336 | anxa5b | -0.175 |
| rpa2 | 0.326 | gstp1 | -0.175 |
| zgc:110540 | 0.315 | cldnh | -0.171 |
| mcm7 | 0.310 | gapdhs | -0.159 |
| slbp | 0.309 | vamp2 | -0.157 |
| fen1 | 0.308 | rtn1a | -0.155 |
| rrm2 | 0.306 | tmem59 | -0.155 |
| mcm4 | 0.305 | CR383676.1 | -0.155 |
| orc4 | 0.304 | rnasekb | -0.152 |
| chaf1a | 0.304 | selenow2b | -0.151 |
| selenoh | 0.300 | fxyd1 | -0.147 |
| rrm1 | 0.299 | calm1a | -0.145 |
| rpa3 | 0.299 | atp6v1e1b | -0.145 |
| nasp | 0.299 | bik | -0.144 |
| stmn1a | 0.297 | gabarapa | -0.144 |
| rfc4 | 0.290 | atp6v0cb | -0.144 |
| lig1 | 0.285 | pvalb5 | -0.143 |
| asf1ba | 0.282 | si:dkeyp-75h12.5 | -0.142 |
| mcm6 | 0.281 | chga | -0.138 |
| dhfr | 0.279 | atp6v1g1 | -0.138 |
| chek1 | 0.278 | wu:fb18f06 | -0.136 |
| esco2 | 0.274 | zgc:65894 | -0.134 |
| prim2 | 0.273 | gnb1a | -0.134 |
| sp8b | 0.271 | atf5a | -0.131 |
| CABZ01005379.1 | 0.269 | selenow1 | -0.131 |
| mcm5 | 0.268 | clstn1 | -0.130 |
| tk1 | 0.267 | si:ch211-153b23.5 | -0.130 |
| rnaseh2a | 0.267 | scg2b | -0.129 |
| mis12 | 0.266 | adcyap1b | -0.129 |
| suv39h1b | 0.266 | nfe2l2a | -0.129 |
| si:ch211-156b7.4 | 0.264 | gng3 | -0.128 |
| cks1b | 0.264 | gsna | -0.128 |
| cdca7b | 0.264 | sncb | -0.128 |
| sumo3b | 0.264 | olfm1b | -0.127 |