DNA cross-link repair 1B
ZFIN 
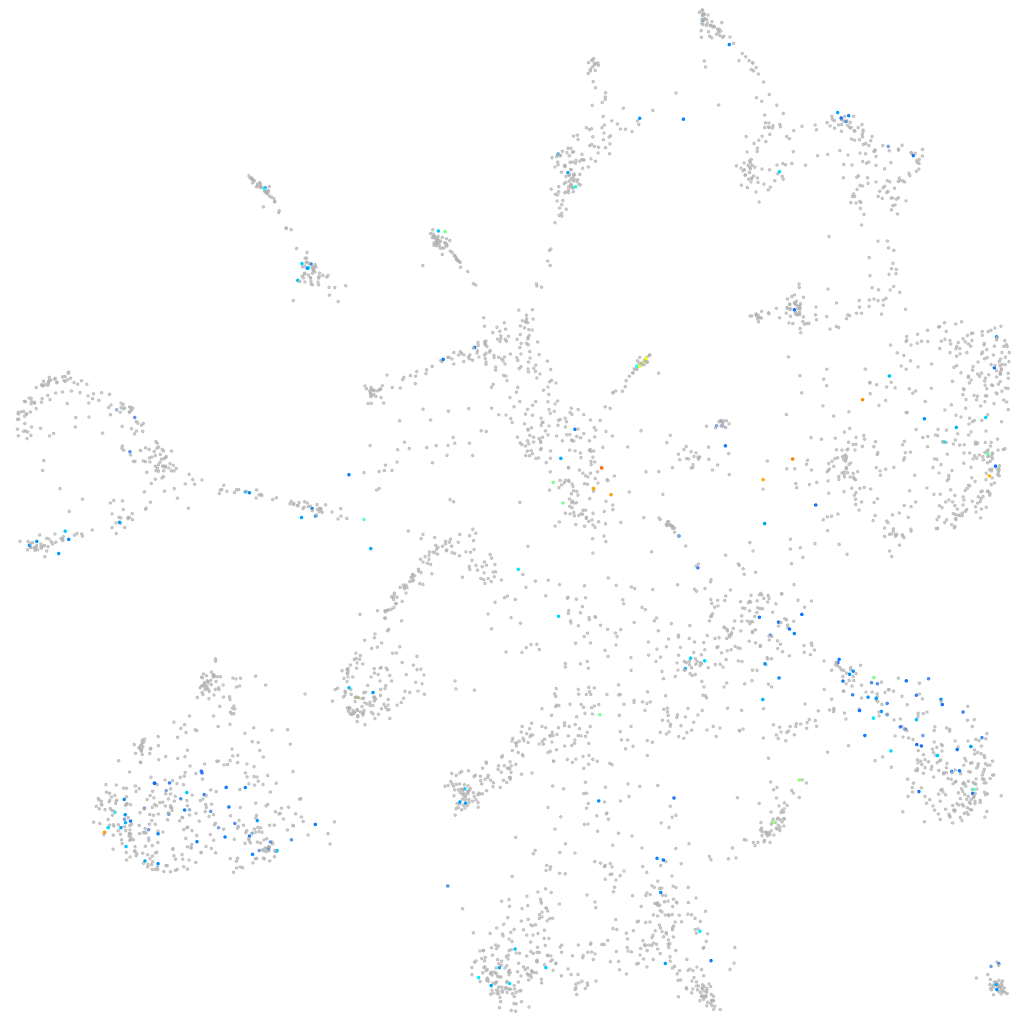
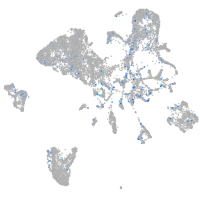
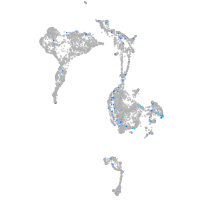
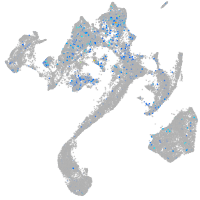
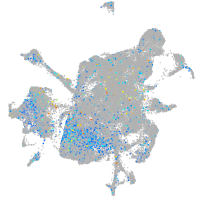
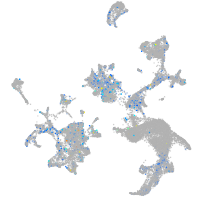
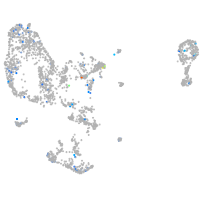
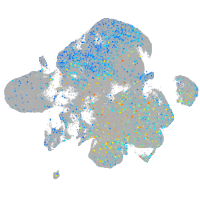
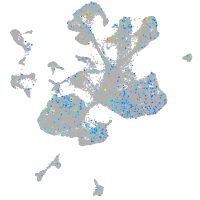
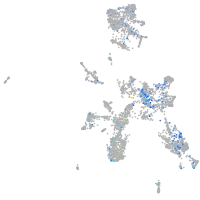
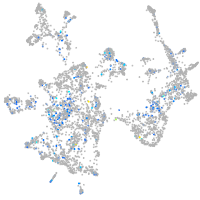
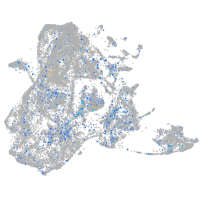
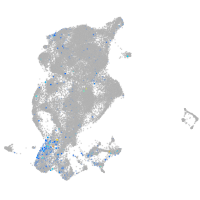
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| CR388132.1 | 0.172 | krt8 | -0.054 |
| BX088525.2 | 0.162 | flna | -0.052 |
| cdh27 | 0.156 | ttn.1 | -0.046 |
| b3glctb | 0.155 | ppp1r12a | -0.045 |
| prkag2b | 0.150 | rpl34 | -0.044 |
| tph2 | 0.149 | ckba | -0.044 |
| slc7a7 | 0.140 | st5 | -0.043 |
| mymk | 0.133 | lxn | -0.042 |
| si:ch1073-228h2.2 | 0.132 | cavin2b | -0.042 |
| BX936461.1 | 0.128 | rps29 | -0.040 |
| neurl1ab | 0.127 | slc7a1 | -0.040 |
| bmp1b | 0.122 | pmp22b | -0.040 |
| rfx4 | 0.121 | luzp1 | -0.038 |
| gabrr2b | 0.118 | rpl36a | -0.038 |
| pcna | 0.117 | rpl26 | -0.038 |
| uhrf1 | 0.117 | slc43a2a | -0.037 |
| LOC103908936 | 0.117 | s100v2 | -0.037 |
| camk1gb | 0.116 | MYADM | -0.037 |
| guca1c | 0.116 | acta1a | -0.037 |
| rnaseh2a | 0.116 | zgc:195173 | -0.037 |
| shisa9b | 0.115 | itgb1a | -0.037 |
| chek1 | 0.115 | copz2 | -0.036 |
| lcp1 | 0.113 | rack1 | -0.036 |
| LOC108190067 | 0.112 | CABZ01058721.1 | -0.036 |
| vtcn1 | 0.112 | adamts8a | -0.036 |
| nasp | 0.110 | gucy1a1 | -0.036 |
| rpa2 | 0.110 | CABZ01092746.1 | -0.036 |
| asb14a | 0.109 | enah | -0.036 |
| kif15 | 0.109 | si:ch73-173h19.3 | -0.035 |
| kcnc1a | 0.108 | mink1 | -0.035 |
| tapbp.2 | 0.108 | rps3 | -0.035 |
| suv39h1b | 0.107 | XLOC-005350 | -0.035 |
| XLOC-043741 | 0.107 | ptprk | -0.035 |
| mibp | 0.106 | irs1 | -0.034 |
| FP016213.1 | 0.105 | ecm1b | -0.033 |