"DNA cross-link repair 1A (PSO2 homolog, S. cerevisiae)"
ZFIN 
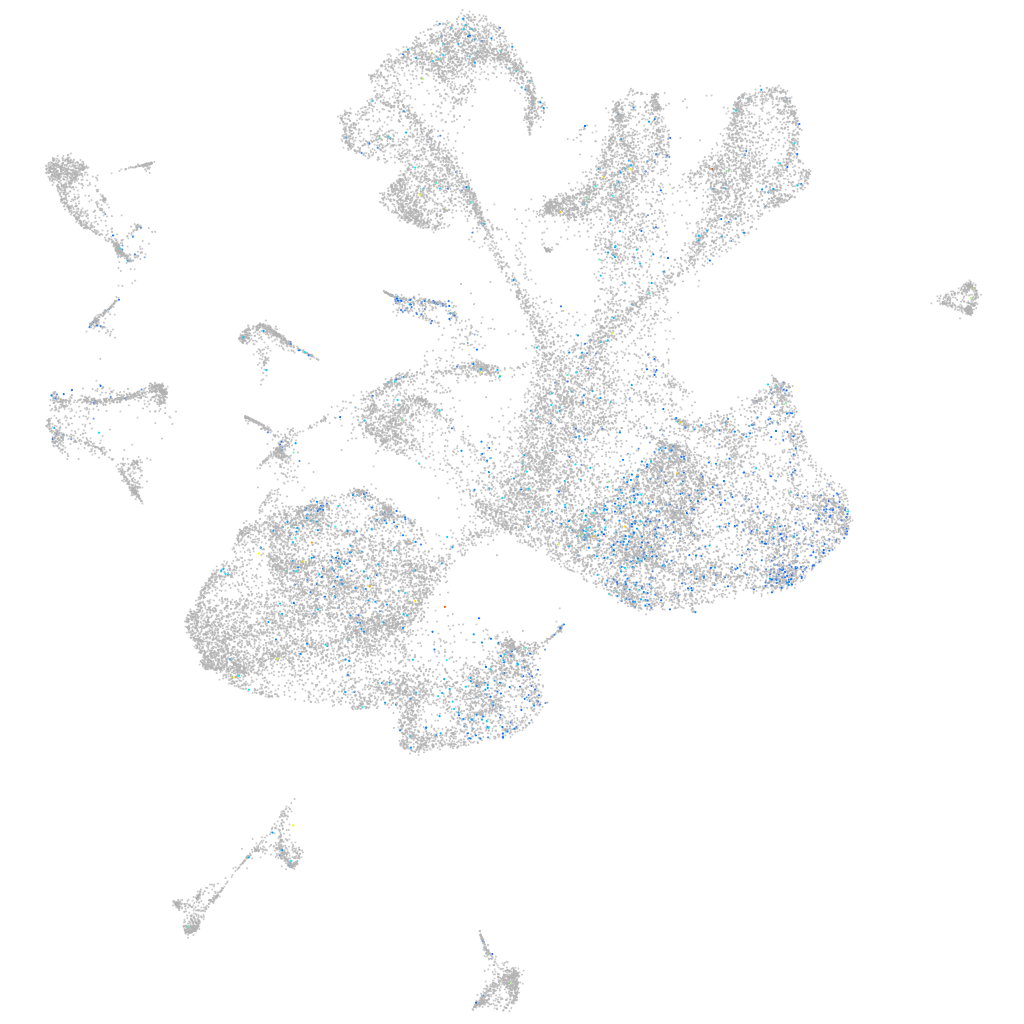
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| pcna | 0.129 | ckbb | -0.056 |
| chaf1a | 0.126 | CR383676.1 | -0.048 |
| rrm2 | 0.124 | ppdpfb | -0.048 |
| zgc:110540 | 0.123 | rtn1a | -0.047 |
| mibp | 0.121 | slc1a2b | -0.045 |
| dut | 0.121 | cotl1 | -0.044 |
| rrm1 | 0.117 | gpm6ab | -0.043 |
| nasp | 0.112 | si:dkeyp-75h12.5 | -0.042 |
| slbp | 0.110 | acbd7 | -0.041 |
| stmn1a | 0.107 | aldocb | -0.041 |
| rpa2 | 0.107 | efhd1 | -0.040 |
| esco2 | 0.107 | rnasekb | -0.040 |
| rpa3 | 0.107 | slc6a1b | -0.039 |
| CABZ01005379.1 | 0.107 | ndrg3a | -0.039 |
| fen1 | 0.106 | gapdhs | -0.039 |
| dek | 0.105 | cx43 | -0.039 |
| banf1 | 0.104 | glula | -0.039 |
| fbxo5 | 0.102 | tob1b | -0.039 |
| tuba8l4 | 0.099 | syt11a | -0.038 |
| cks1b | 0.099 | h1f0 | -0.038 |
| zgc:165555.3 | 0.099 | fez1 | -0.037 |
| rfc3 | 0.098 | pvalb2 | -0.037 |
| nutf2l | 0.097 | si:ch211-260e23.9 | -0.037 |
| lbr | 0.097 | smox | -0.037 |
| zgc:110216 | 0.097 | cadm4 | -0.036 |
| suv39h1b | 0.096 | calm1a | -0.036 |
| ccne2 | 0.096 | gpr37l1b | -0.036 |
| unga | 0.095 | slc4a4a | -0.035 |
| prim2 | 0.095 | marcksl1b | -0.035 |
| lig1 | 0.095 | itm2ba | -0.035 |
| mcm7 | 0.095 | elavl3 | -0.035 |
| ssrp1a | 0.094 | gpm6aa | -0.035 |
| anp32b | 0.094 | elavl4 | -0.034 |
| mis12 | 0.094 | ptn | -0.034 |
| rnaseh2a | 0.093 | sesn1 | -0.034 |