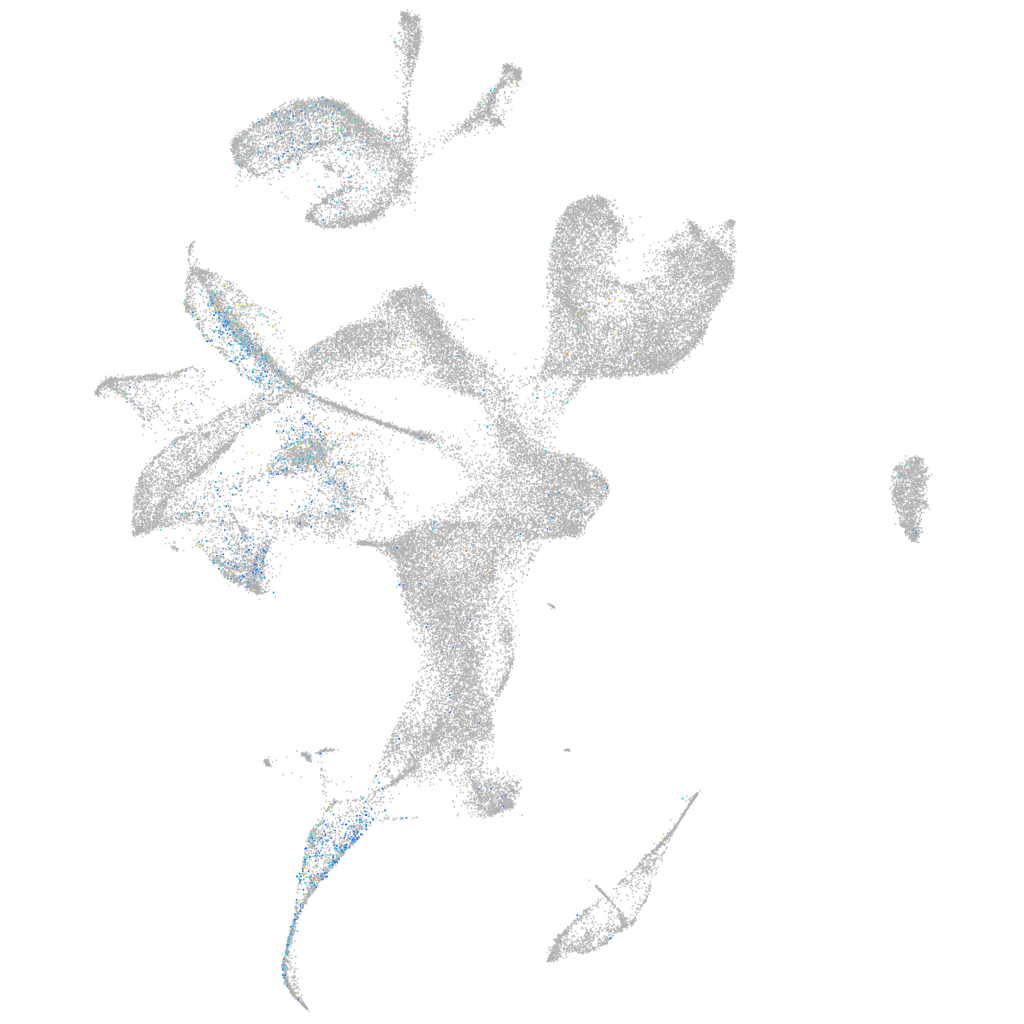
dachsous cadherin-related 1b
ZFIN 
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| vat1 | 0.165 | hmgb2a | -0.087 |
| dct | 0.165 | fabp7a | -0.073 |
| pmelb | 0.163 | anp32e | -0.068 |
| slc45a2 | 0.160 | ptmab | -0.056 |
| tyrp1a | 0.160 | neurod4 | -0.055 |
| tyrp1b | 0.160 | si:ch73-1a9.3 | -0.051 |
| pmela | 0.160 | si:ch73-281n10.2 | -0.050 |
| gpr143 | 0.159 | syt5b | -0.048 |
| oca2 | 0.158 | crx | -0.048 |
| slc24a5 | 0.155 | histh1l | -0.047 |
| tyr | 0.155 | rrm1 | -0.047 |
| rab38 | 0.155 | h2afva | -0.047 |
| agtrap | 0.150 | stmn1a | -0.047 |
| tspan36 | 0.149 | mki67 | -0.046 |
| qdpra | 0.147 | dek | -0.046 |
| tspan10 | 0.144 | pcna | -0.045 |
| mitfa | 0.139 | rrm2 | -0.045 |
| cracr2ab | 0.139 | anp32b | -0.045 |
| pah | 0.137 | vsx1 | -0.044 |
| zgc:110591 | 0.137 | ccnd1 | -0.044 |
| tmsb | 0.136 | XLOC-042899 | -0.044 |
| sytl2b | 0.134 | zgc:110540 | -0.044 |
| bace2 | 0.134 | ccna2 | -0.044 |
| tmem88b | 0.132 | dut | -0.043 |
| hps1 | 0.131 | nasp | -0.043 |
| LOC103910009 | 0.131 | chaf1a | -0.043 |
| rab32a | 0.130 | hmga1a | -0.042 |
| FP085398.1 | 0.129 | fbxo5 | -0.042 |
| fabp11b | 0.128 | fzd5 | -0.042 |
| col9a2 | 0.125 | nutf2l | -0.041 |
| triobpa | 0.125 | zgc:110216 | -0.041 |
| islr2 | 0.125 | lig1 | -0.041 |
| sparc | 0.124 | CABZ01005379.1 | -0.041 |
| col4a5 | 0.124 | slbp | -0.040 |
| col4a6 | 0.124 | vsx2 | -0.040 |