death-associated protein
ZFIN 
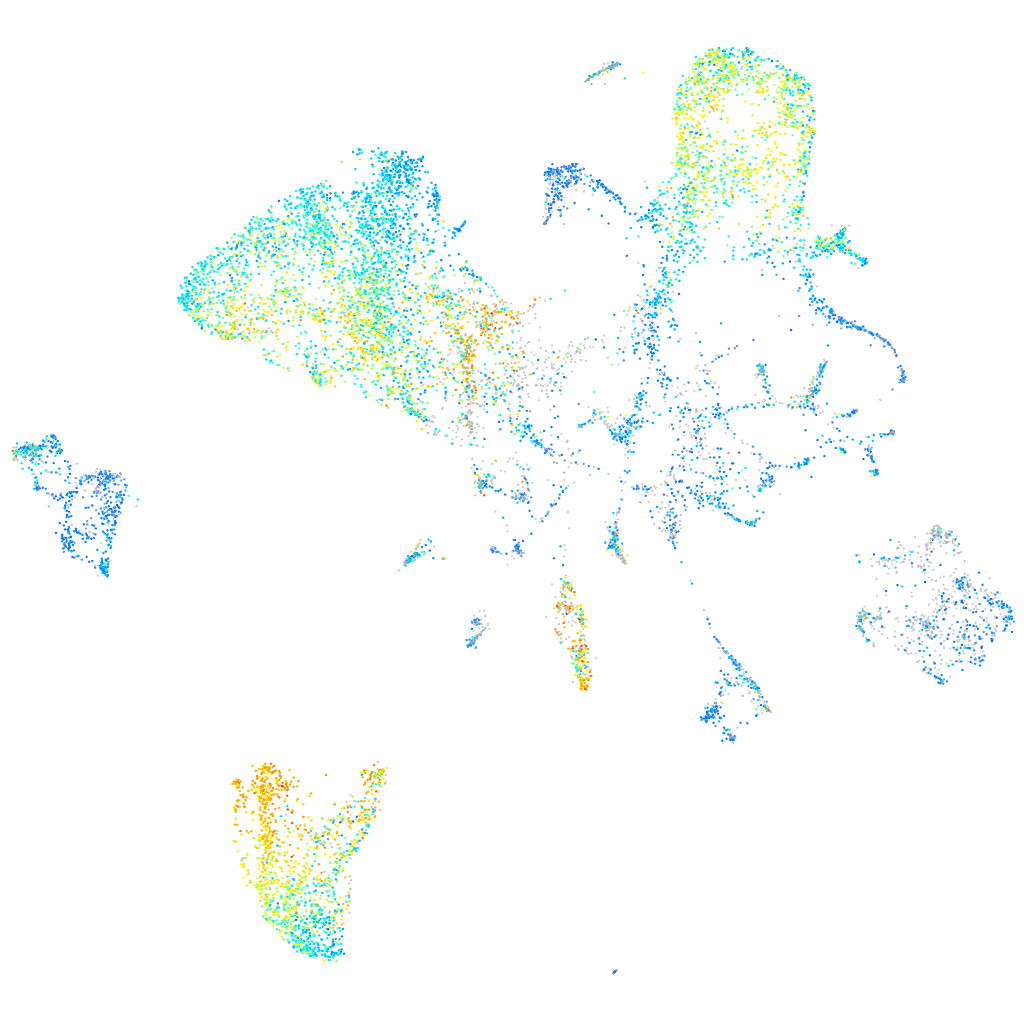
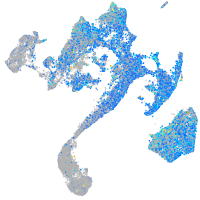
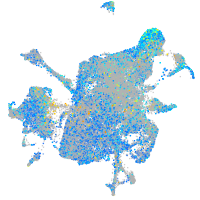
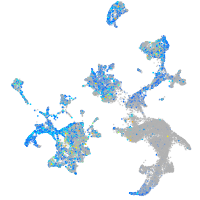
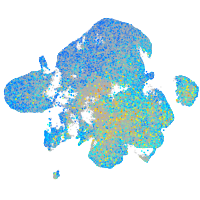
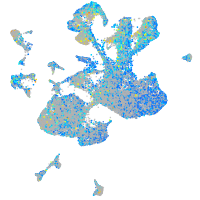
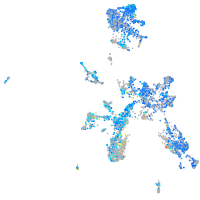
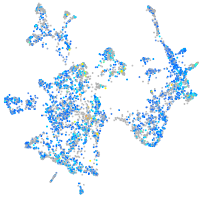
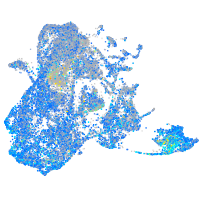
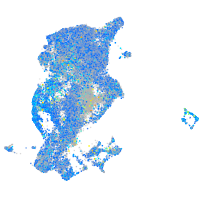
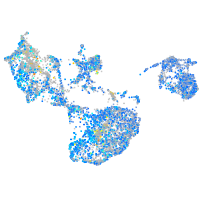
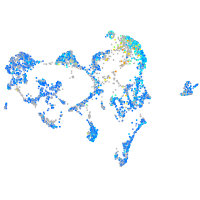
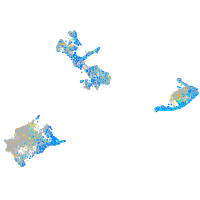
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| gatm | 0.592 | si:ch211-222l21.1 | -0.474 |
| gapdh | 0.561 | si:ch73-1a9.3 | -0.437 |
| gamt | 0.556 | hmgn6 | -0.437 |
| ahcy | 0.551 | h3f3d | -0.431 |
| nupr1b | 0.539 | hmgb3a | -0.428 |
| mat1a | 0.525 | marcksl1b | -0.402 |
| glud1b | 0.504 | epcam | -0.394 |
| fbp1b | 0.497 | hmgb2a | -0.389 |
| slc16a10 | 0.474 | nucks1a | -0.388 |
| cx32.3 | 0.466 | marcksb | -0.385 |
| suclg1 | 0.466 | id1 | -0.385 |
| zgc:92744 | 0.465 | hnrnpaba | -0.384 |
| aldh7a1 | 0.465 | hmgb1b | -0.384 |
| aqp12 | 0.461 | hmgb1a | -0.382 |
| bhmt | 0.457 | aldoaa | -0.380 |
| scp2a | 0.450 | fthl27 | -0.378 |
| mcfd2 | 0.442 | si:ch73-281n10.2 | -0.377 |
| cdo1 | 0.438 | jpt1b | -0.377 |
| gpx4a | 0.431 | ubc | -0.373 |
| apoa4b.1 | 0.423 | khdrbs1a | -0.373 |
| krtcap2 | 0.421 | syncrip | -0.368 |
| rltgr | 0.417 | seta | -0.365 |
| ssr3 | 0.413 | acin1a | -0.364 |
| sod1 | 0.413 | cx43.4 | -0.359 |
| hdlbpa | 0.412 | h2afvb | -0.359 |
| ssr2 | 0.412 | ywhaz | -0.356 |
| apoc2 | 0.411 | hmgn7 | -0.355 |
| abat | 0.410 | cirbpb | -0.354 |
| ppdpfa | 0.410 | anp32a | -0.353 |
| sult2st2 | 0.410 | hmgn2 | -0.351 |
| atp5if1b | 0.409 | ptmab | -0.349 |
| cox6a1 | 0.409 | hdac1 | -0.348 |
| eno3 | 0.408 | tuba8l4 | -0.347 |
| suclg2 | 0.408 | cd9b | -0.343 |
| mdh1aa | 0.408 | bcam | -0.342 |