dishevelled associated activator of morphogenesis 1a
ZFIN 
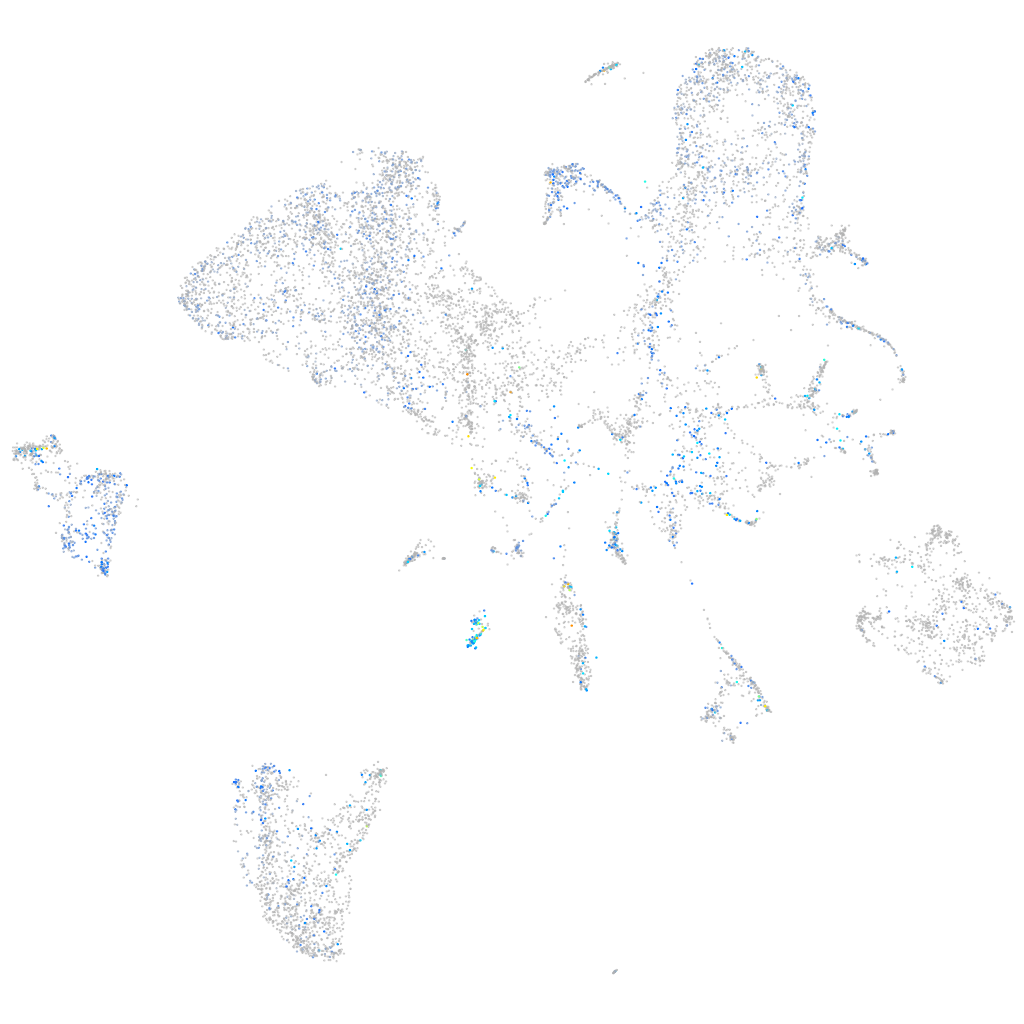
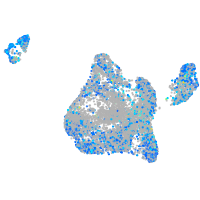
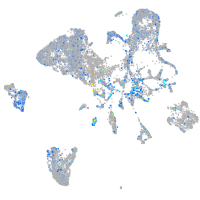
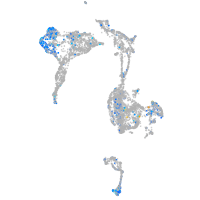
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| si:dkey-96g2.1 | 0.254 | apoc1 | -0.110 |
| zgc:113142 | 0.249 | gamt | -0.100 |
| blnk | 0.216 | gatm | -0.090 |
| zgc:193726 | 0.213 | si:dkey-16p21.8 | -0.089 |
| cldn2 | 0.205 | fabp10a | -0.085 |
| cdh1 | 0.198 | agxtb | -0.084 |
| sox9b | 0.194 | rbp2b | -0.083 |
| cobll1a | 0.189 | bhmt | -0.083 |
| cx30.3 | 0.187 | hao1 | -0.079 |
| clic2 | 0.187 | serpina1l | -0.079 |
| pdzk1ip1 | 0.181 | apoa1b | -0.079 |
| dab2ipb | 0.174 | uox | -0.079 |
| cd9b | 0.173 | apom | -0.078 |
| slc4a4b | 0.172 | gapdh | -0.077 |
| si:dkey-163m14.2 | 0.166 | tdo2a | -0.077 |
| epcam | 0.165 | serpina1 | -0.076 |
| btk | 0.164 | apoa2 | -0.076 |
| onecut1 | 0.163 | zgc:123103 | -0.076 |
| nectin4b | 0.162 | apoa4b.1 | -0.076 |
| rassf7b | 0.159 | mat1a | -0.075 |
| phactr4a | 0.159 | gc | -0.075 |
| b3gnt2l | 0.159 | ces2 | -0.075 |
| GUCY1A2 | 0.155 | ttc36 | -0.074 |
| enc1 | 0.155 | ttr | -0.074 |
| foxp4 | 0.155 | grhprb | -0.074 |
| CR339041.2 | 0.154 | apoc2 | -0.073 |
| id2a | 0.153 | ftcd | -0.073 |
| hmga2 | 0.152 | upb1 | -0.073 |
| CR383676.1 | 0.150 | fetub | -0.072 |
| zgc:198329 | 0.149 | pnp4b | -0.071 |
| CR396595.1 | 0.149 | ahcy | -0.071 |
| plecb | 0.149 | scp2a | -0.071 |
| gmnn | 0.148 | LOC110437731 | -0.071 |
| jun | 0.148 | rbp4 | -0.071 |
| f11r.1 | 0.148 | hgd | -0.071 |