cytoplasmic FMR1 interacting protein 2
ZFIN 
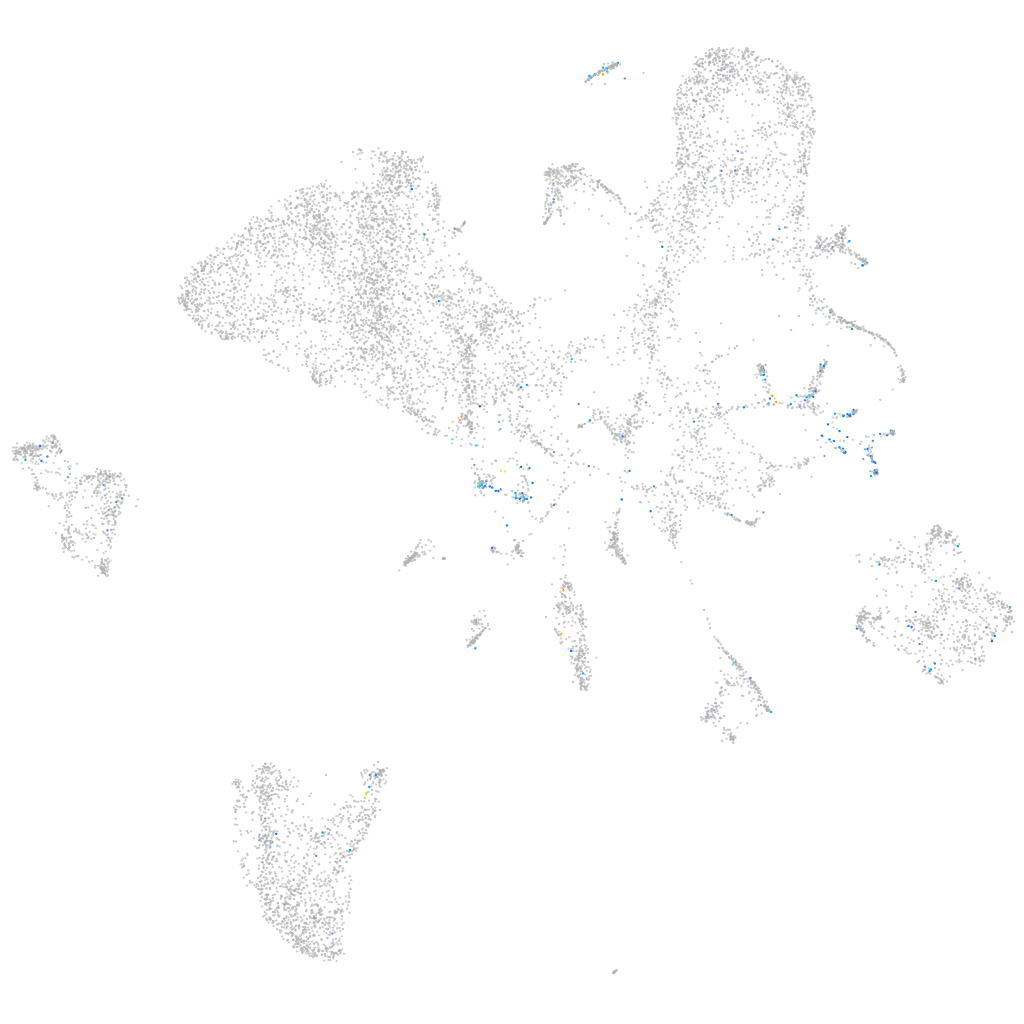
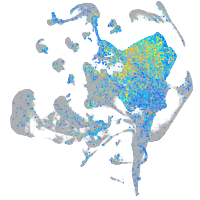
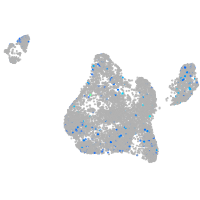
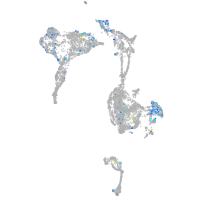
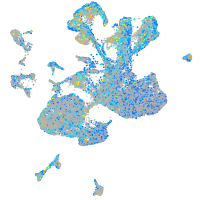
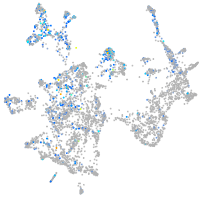
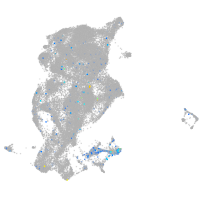
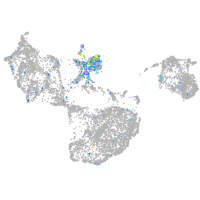
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| scg3 | 0.217 | gapdh | -0.110 |
| pax6b | 0.212 | ahcy | -0.106 |
| neurod1 | 0.211 | aldob | -0.104 |
| scg2a | 0.209 | gamt | -0.100 |
| stxbp1a | 0.207 | fbp1b | -0.095 |
| pcsk1nl | 0.205 | aldh6a1 | -0.095 |
| vat1 | 0.204 | mat1a | -0.093 |
| gpc1a | 0.201 | gstt1a | -0.090 |
| vamp2 | 0.201 | scp2a | -0.089 |
| elavl3 | 0.200 | eno3 | -0.087 |
| hcn4l | 0.195 | gatm | -0.086 |
| tuba1c | 0.193 | suclg1 | -0.084 |
| gng3 | 0.193 | mgst1.2 | -0.084 |
| dmtn | 0.191 | nupr1b | -0.083 |
| pcsk1 | 0.189 | gstr | -0.082 |
| cplx2 | 0.189 | cx32.3 | -0.082 |
| si:ch73-287m6.1 | 0.188 | eef1da | -0.081 |
| mir7a-1 | 0.188 | agxtb | -0.080 |
| atp1a3a | 0.187 | aldh7a1 | -0.080 |
| arf3b | 0.187 | gnmt | -0.080 |
| LOC100537384 | 0.186 | agxta | -0.079 |
| gdi1 | 0.186 | dap | -0.079 |
| lysmd2 | 0.185 | pnp4b | -0.077 |
| rnasekb | 0.185 | aldh1l1 | -0.077 |
| id4 | 0.184 | cebpd | -0.077 |
| atp6v0cb | 0.184 | fabp10a | -0.077 |
| scgn | 0.183 | upb1 | -0.077 |
| dpysl5a | 0.182 | glud1b | -0.077 |
| tmsb | 0.182 | fdx1 | -0.076 |
| jagn1a | 0.181 | phyhd1 | -0.076 |
| syt1a | 0.180 | aldh9a1a.1 | -0.076 |
| cntnap1 | 0.180 | uox | -0.076 |
| myt1b | 0.180 | ttc36 | -0.075 |
| cnrip1a | 0.179 | suclg2 | -0.075 |
| rprmb | 0.179 | nipsnap3a | -0.075 |