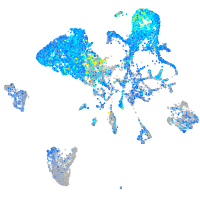
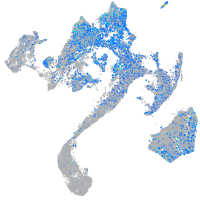
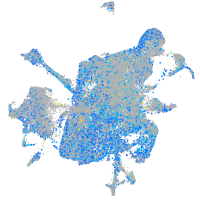
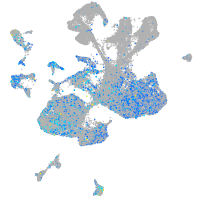
cytochrome b5 type A (microsomal)
ZFIN 
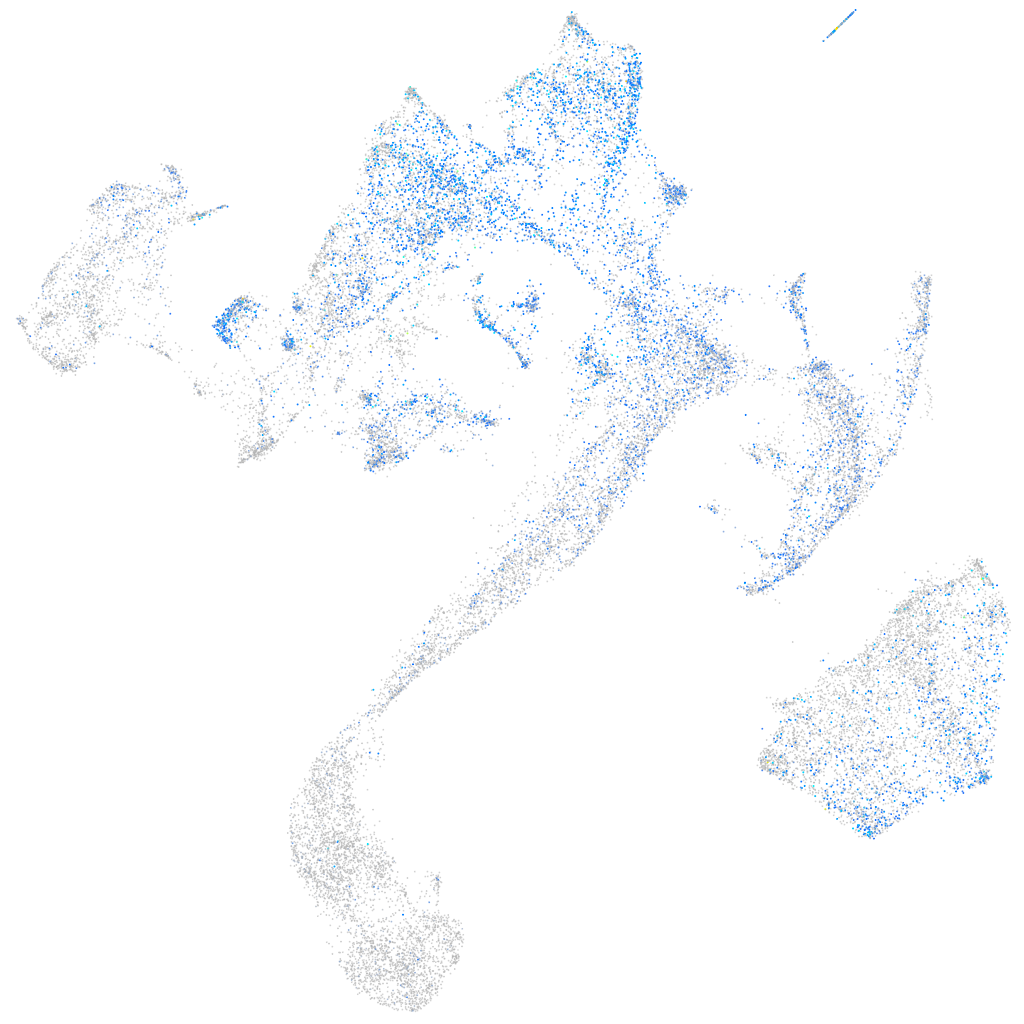
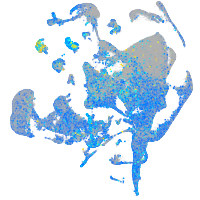
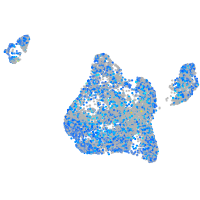
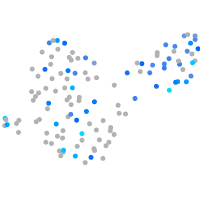
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| tpm4a | 0.308 | ttn.1 | -0.226 |
| actb2 | 0.302 | atp2a1 | -0.224 |
| actb1 | 0.290 | ckmb | -0.219 |
| marcksl1a | 0.286 | ak1 | -0.218 |
| zgc:153867 | 0.283 | aldoab | -0.217 |
| pdia3 | 0.276 | tnnc2 | -0.217 |
| krt18a.1 | 0.273 | actc1b | -0.216 |
| pmp22a | 0.269 | tmem38a | -0.215 |
| mfap2 | 0.265 | ckma | -0.215 |
| ppib | 0.262 | neb | -0.214 |
| cfl1 | 0.262 | ttn.2 | -0.212 |
| tmsb4x | 0.261 | gapdh | -0.206 |
| ssr3 | 0.257 | si:ch73-367p23.2 | -0.205 |
| tmem258 | 0.253 | mybphb | -0.202 |
| calr | 0.246 | ldb3b | -0.201 |
| kdelr2b | 0.245 | actn3b | -0.200 |
| h3f3a | 0.245 | tpma | -0.199 |
| fkbp14 | 0.243 | actn3a | -0.199 |
| ssr4 | 0.243 | acta1b | -0.198 |
| krt8 | 0.241 | mylpfa | -0.197 |
| zgc:92744 | 0.237 | ank1a | -0.196 |
| lasp1 | 0.236 | srl | -0.195 |
| hmgn2 | 0.236 | hhatla | -0.195 |
| zgc:153675 | 0.235 | myl1 | -0.194 |
| lrrc17 | 0.235 | CABZ01078594.1 | -0.194 |
| serp1 | 0.234 | myom1a | -0.194 |
| sec61g | 0.231 | ldb3a | -0.194 |
| cnn2 | 0.231 | tnnt3a | -0.193 |
| calua | 0.229 | smyd1a | -0.192 |
| xbp1 | 0.229 | cav3 | -0.192 |
| twist1a | 0.228 | mylz3 | -0.190 |
| col5a2a | 0.228 | si:ch211-266g18.10 | -0.189 |
| ssr2 | 0.228 | pabpc4 | -0.189 |
| rbp5 | 0.227 | tmod4 | -0.188 |
| col5a1 | 0.227 | desma | -0.187 |