connexin 32.2
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature 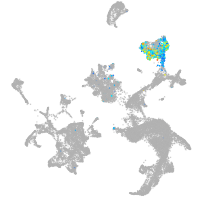 |
pronephros 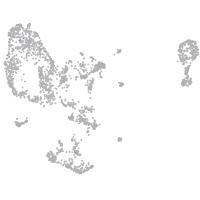 |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| cmklr1 | 0.731 | hbae1.1 | -0.222 |
| havcr1 | 0.729 | hbae1.3 | -0.210 |
| mpeg1.1 | 0.717 | hbae3 | -0.209 |
| mfap4 | 0.704 | hbbe2 | -0.208 |
| si:dkey-5n18.1 | 0.702 | hbbe1.3 | -0.207 |
| ctss2.2 | 0.677 | hbbe1.1 | -0.201 |
| tnfb | 0.664 | fth1a | -0.191 |
| si:zfos-1069f5.1 | 0.653 | cahz | -0.191 |
| lgals2a | 0.653 | hemgn | -0.180 |
| slc7a7 | 0.646 | zgc:56095 | -0.180 |
| lgals3bpb | 0.637 | hbbe1.2 | -0.180 |
| ccl34a.4 | 0.618 | lmo2 | -0.177 |
| si:ch211-147m6.1 | 0.615 | alas2 | -0.168 |
| si:ch211-194m7.3 | 0.610 | si:ch211-250g4.3 | -0.167 |
| lygl1 | 0.608 | slc4a1a | -0.158 |
| nfkbiaa | 0.601 | nt5c2l1 | -0.154 |
| tspan36 | 0.597 | creg1 | -0.154 |
| ms4a17a.10 | 0.593 | blvrb | -0.150 |
| CU499330.1 | 0.591 | zgc:163057 | -0.149 |
| si:ch211-212k18.7 | 0.588 | epb41b | -0.148 |
| slc3a2a | 0.585 | si:ch211-207c6.2 | -0.143 |
| ctsl.1 | 0.580 | tspo | -0.136 |
| stoml3b | 0.579 | nmt1b | -0.135 |
| ctsc | 0.577 | aqp1a.1 | -0.135 |
| ccl34b.1 | 0.576 | plac8l1 | -0.131 |
| acod1 | 0.575 | tmod4 | -0.130 |
| ctsba | 0.571 | arl4aa | -0.124 |
| tnfsf12 | 0.569 | histh1l | -0.121 |
| marco | 0.558 | znfl2a | -0.121 |
| cd83 | 0.556 | hbae5 | -0.120 |
| gm2a | 0.554 | sptb | -0.119 |
| c1qc | 0.549 | uros | -0.119 |
| itgae.1 | 0.546 | si:ch211-227m13.1 | -0.119 |
| fcer1gl | 0.542 | tfr1a | -0.117 |
| irf8 | 0.539 | si:ch211-222l21.1 | -0.116 |




