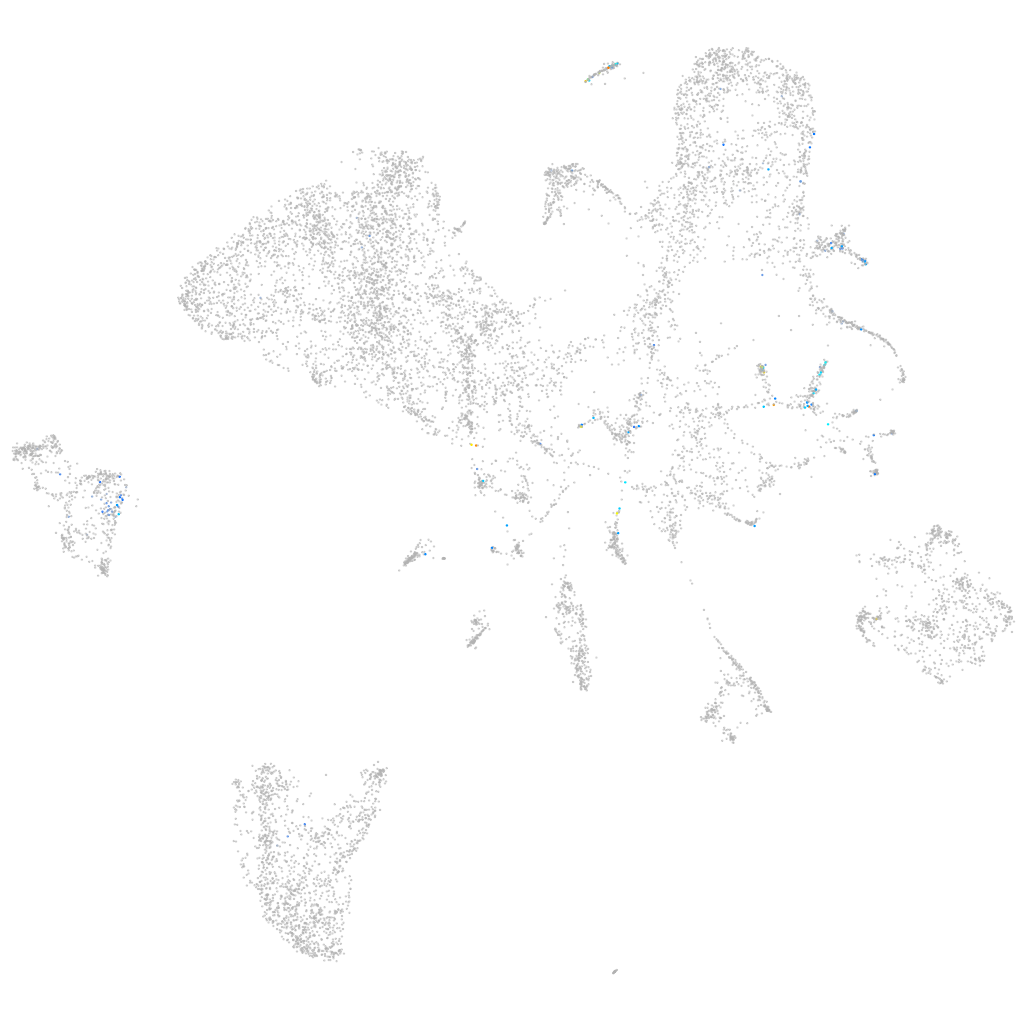
connexin 28.8
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros 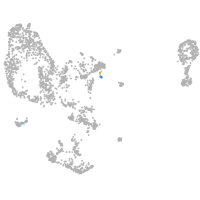 |
neural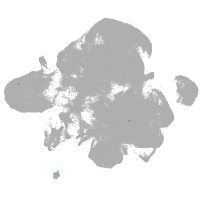 |
spinal cord / glia |
eye |
taste / olfactory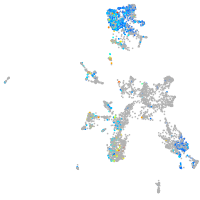 |
otic / lateral line |
epidermis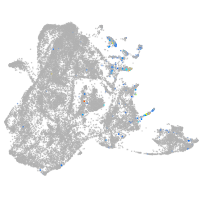 |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| XLOC-011873 | 0.225 | gamt | -0.085 |
| insm1b | 0.220 | ahcy | -0.079 |
| hepacam2 | 0.211 | gapdh | -0.078 |
| scg3 | 0.207 | bhmt | -0.072 |
| RBP1 (1 of many) | 0.206 | gatm | -0.071 |
| zgc:101731 | 0.205 | mat1a | -0.070 |
| egr4 | 0.204 | nupr1b | -0.069 |
| si:dkey-224b4.5 | 0.193 | aldh7a1 | -0.068 |
| zgc:194908 | 0.188 | apoa4b.1 | -0.067 |
| slc35g2a | 0.188 | fabp3 | -0.067 |
| capsla | 0.187 | apoc2 | -0.066 |
| arhgef3 | 0.185 | apoa1b | -0.066 |
| fev | 0.181 | apoc1 | -0.065 |
| si:dkeyp-72e1.7 | 0.179 | aqp12 | -0.064 |
| dll4 | 0.177 | afp4 | -0.063 |
| id4 | 0.176 | fbp1b | -0.063 |
| calm1b | 0.174 | si:dkey-16p21.8 | -0.062 |
| cfap43 | 0.174 | abat | -0.061 |
| rgs16 | 0.173 | hdlbpa | -0.061 |
| si:dkey-83f18.10 | 0.173 | gstt1a | -0.061 |
| ret | 0.171 | agxta | -0.060 |
| dusp2 | 0.166 | agxtb | -0.060 |
| syt1a | 0.165 | tfa | -0.059 |
| cldn11a | 0.163 | apoa2 | -0.059 |
| bdnf | 0.163 | shmt1 | -0.058 |
| zgc:162928 | 0.163 | cx32.3 | -0.058 |
| micu3b | 0.163 | gcshb | -0.058 |
| si:dkey-83f18.8 | 0.162 | gstr | -0.058 |
| si:dkey-28d5.9 | 0.162 | gnmt | -0.057 |
| tpm1 | 0.162 | apobb.1 | -0.057 |
| neurod1 | 0.162 | ftcd | -0.057 |
| si:ch211-212k5.3 | 0.161 | scp2a | -0.056 |
| pcsk1 | 0.161 | serpina1l | -0.056 |
| si:ch211-63p21.4 | 0.161 | hspd1 | -0.056 |
| prf1.7 | 0.161 | nipsnap3a | -0.055 |