cut-like homeobox 1a
ZFIN 
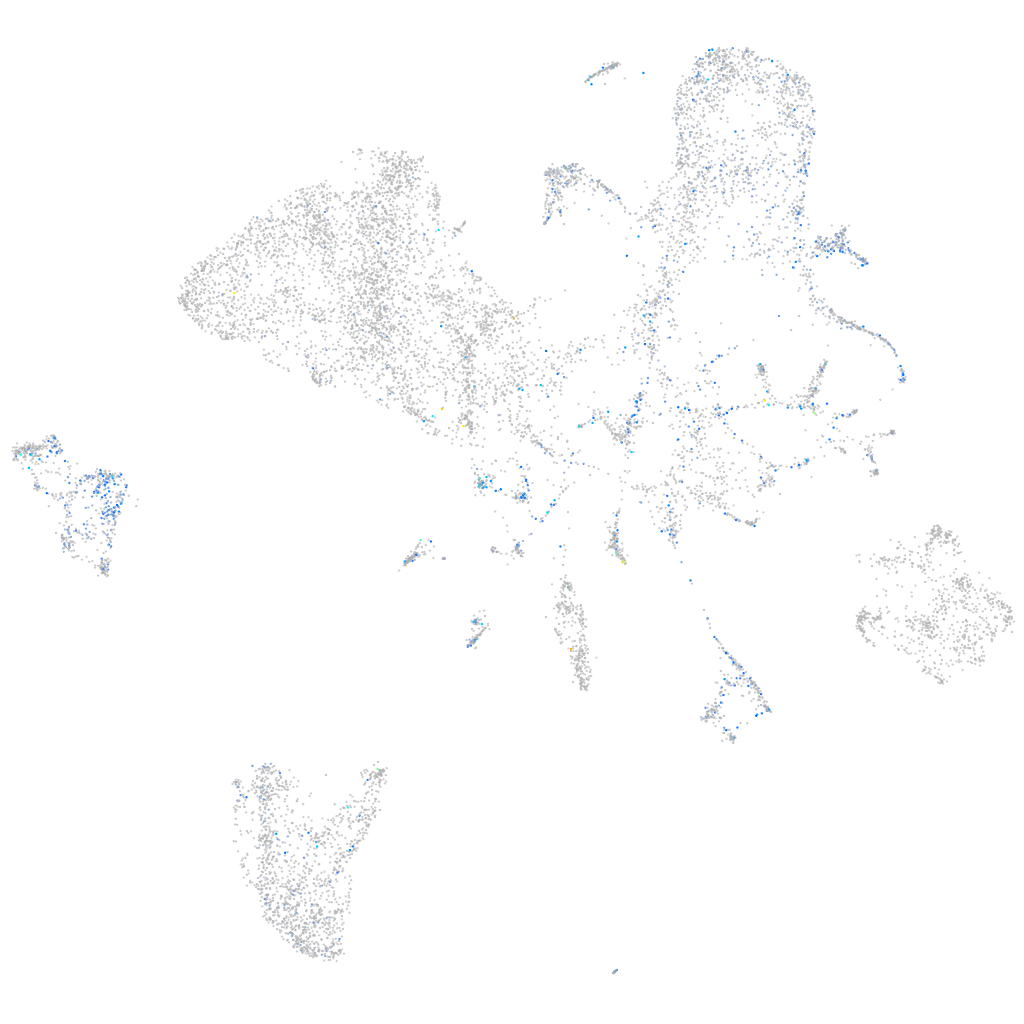
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| epcam | 0.193 | gamt | -0.169 |
| tmsb4x | 0.179 | bhmt | -0.164 |
| pfn1 | 0.173 | apoc1 | -0.162 |
| hnrnpa0l | 0.172 | agxtb | -0.151 |
| ier2b | 0.172 | gatm | -0.150 |
| CR383676.1 | 0.170 | mat1a | -0.149 |
| phlda2 | 0.168 | rbp2b | -0.147 |
| h3f3a | 0.167 | fabp10a | -0.144 |
| ddit3 | 0.165 | aqp12 | -0.143 |
| mcl1a | 0.163 | uox | -0.142 |
| ptmaa | 0.161 | serpina1l | -0.142 |
| atf4b | 0.161 | zgc:123103 | -0.142 |
| si:ch1073-429i10.3.1 | 0.161 | apoa1b | -0.142 |
| cldnb | 0.159 | apoa2 | -0.142 |
| ubc | 0.158 | grhprb | -0.142 |
| hbegfa | 0.157 | ttc36 | -0.141 |
| gnb1a | 0.156 | serpina1 | -0.141 |
| btg1 | 0.155 | tfa | -0.141 |
| actb1 | 0.155 | fgg | -0.139 |
| ccni | 0.155 | hpda | -0.138 |
| spint2 | 0.155 | fgb | -0.138 |
| pfn2 | 0.155 | apom | -0.138 |
| cap1 | 0.154 | ttr | -0.138 |
| midn | 0.154 | LOC110437731 | -0.137 |
| kdm6bb | 0.153 | pygl | -0.137 |
| cldn7b | 0.153 | ces2 | -0.137 |
| cfl1l | 0.153 | pnp4b | -0.137 |
| wu:fb55g09 | 0.153 | ambp | -0.137 |
| chmp5b | 0.153 | hao1 | -0.137 |
| elf3 | 0.152 | gapdh | -0.137 |
| tagln2 | 0.151 | fetub | -0.136 |
| gapdhs | 0.150 | f2 | -0.136 |
| ier5 | 0.149 | c9 | -0.136 |
| krt4 | 0.149 | gc | -0.136 |
| twf1a | 0.148 | kng1 | -0.136 |