cullin 3a
ZFIN 
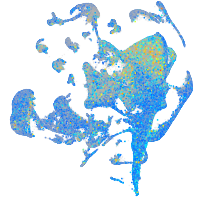
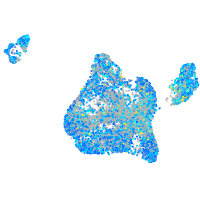
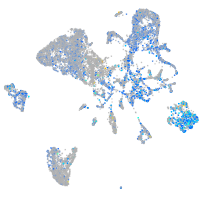
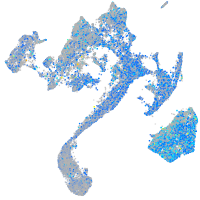
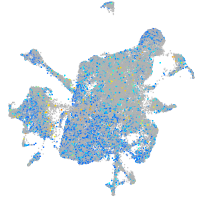
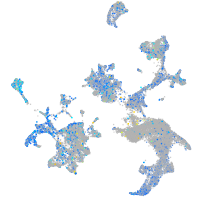
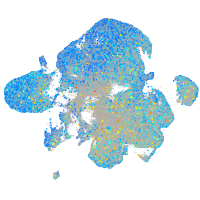
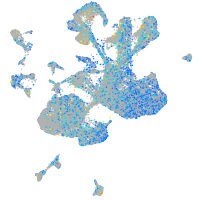
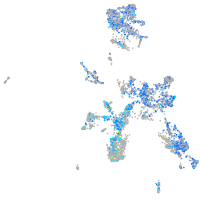
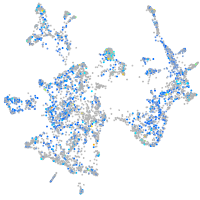
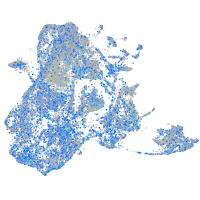
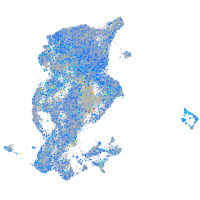
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| hnrnpabb | 0.174 | si:ch211-139a5.9 | -0.147 |
| syncrip | 0.170 | aldob | -0.133 |
| khdrbs1a | 0.169 | grhprb | -0.118 |
| sart1 | 0.165 | gamt | -0.117 |
| hmgb2b | 0.165 | aldh6a1 | -0.116 |
| top1l | 0.165 | gapdh | -0.115 |
| setb | 0.163 | fbp1b | -0.109 |
| hnrnpaba | 0.163 | nit2 | -0.109 |
| si:ch73-281n10.2 | 0.161 | eno3 | -0.109 |
| smc1al | 0.161 | rgrb | -0.106 |
| psme3 | 0.159 | si:dkey-16p21.8 | -0.105 |
| cirbpb | 0.154 | gcshb | -0.104 |
| si:ch211-222l21.1 | 0.154 | suclg1 | -0.104 |
| hdac1 | 0.153 | atp5mc3b | -0.103 |
| hmga1a | 0.153 | gstt1a | -0.103 |
| snrpg | 0.150 | hao1 | -0.103 |
| sap18 | 0.148 | atp1b1a | -0.102 |
| snrpd2 | 0.148 | cdaa | -0.102 |
| erh | 0.148 | atp5l | -0.102 |
| leo1 | 0.147 | pnp6 | -0.099 |
| cbx3a | 0.147 | mdh1aa | -0.099 |
| ncl | 0.147 | atp5f1e | -0.098 |
| hnrnpa0b | 0.147 | prdx2 | -0.098 |
| hmgn6 | 0.147 | glud1b | -0.097 |
| si:ch211-288g17.3 | 0.146 | rps2 | -0.096 |
| ewsr1a | 0.145 | rpl37 | -0.096 |
| ddx39aa | 0.145 | dhdhl | -0.095 |
| ruvbl2 | 0.145 | hoga1 | -0.095 |
| h3f3d | 0.144 | upb1 | -0.095 |
| h2afvb | 0.144 | slc25a5 | -0.093 |
| myef2 | 0.144 | dera | -0.093 |
| marcksb | 0.144 | grxcr1b | -0.093 |
| hmgb2a | 0.144 | atp5fa1 | -0.093 |
| snrpd1 | 0.143 | pbld | -0.093 |
| snrpe | 0.142 | lgmn | -0.093 |