cullin 2
ZFIN 
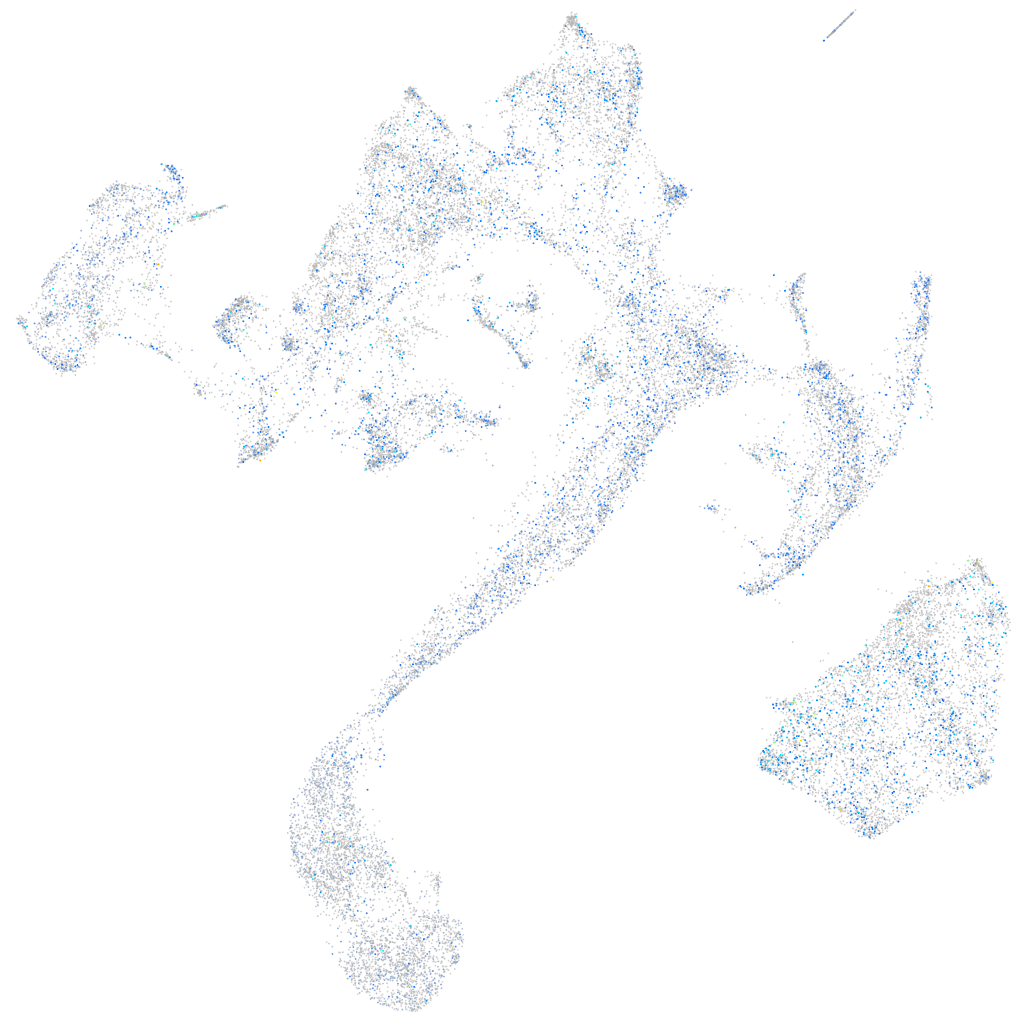
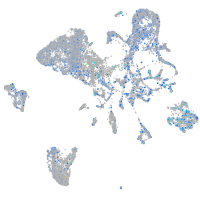
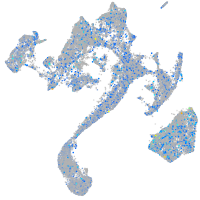
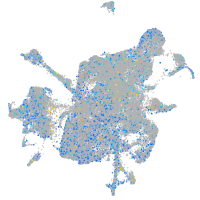
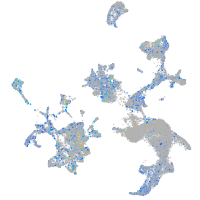
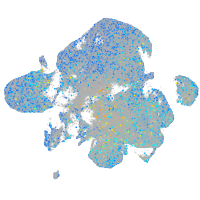
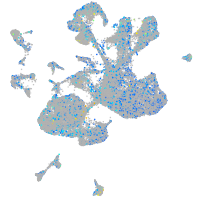
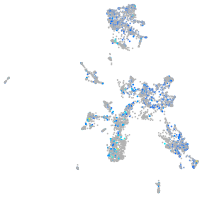
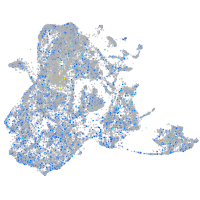
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| ppp2cb | 0.064 | pvalb1 | -0.050 |
| zc3h13 | 0.063 | pvalb2 | -0.048 |
| smarca4a | 0.062 | rpl37 | -0.047 |
| rhoaa | 0.062 | mylpfb | -0.044 |
| hnrnpaba | 0.061 | tnni2a.4 | -0.043 |
| nucks1a | 0.061 | slc25a4 | -0.042 |
| ilf2 | 0.061 | tnnt3b | -0.041 |
| hdac1 | 0.061 | bhmt | -0.040 |
| pdap1a | 0.060 | mylz3 | -0.040 |
| hnrnph1l | 0.060 | rps29 | -0.039 |
| hspb1 | 0.060 | hsc70 | -0.038 |
| marcksb | 0.060 | rbp4 | -0.038 |
| srsf5a | 0.060 | si:ch73-366l1.5 | -0.038 |
| tardbp | 0.060 | myhz1.1 | -0.037 |
| ube2v2 | 0.059 | myoz1b | -0.037 |
| srrm1 | 0.059 | si:ch211-255p10.3 | -0.037 |
| hnrnpa1a | 0.059 | zgc:114188 | -0.037 |
| ddx39ab | 0.059 | CABZ01061524.1 | -0.036 |
| hnrnpabb | 0.059 | ampd1 | -0.035 |
| hmgn6 | 0.059 | rps10 | -0.035 |
| eny2 | 0.059 | si:dkey-16p21.8 | -0.035 |
| hdgfl2 | 0.058 | myom2a | -0.035 |
| hmga1a | 0.058 | mylpfa | -0.034 |
| igf2bp1 | 0.058 | rps17 | -0.033 |
| eif5a2 | 0.058 | ryr3 | -0.033 |
| psmb7 | 0.058 | eef1da | -0.033 |
| zmat2 | 0.058 | myhz1.2 | -0.033 |
| srsf1a | 0.058 | igfn1.3 | -0.033 |
| ppig | 0.058 | atp2a1l | -0.033 |
| snrnp70 | 0.058 | si:ch211-251b21.1 | -0.032 |
| pomp | 0.058 | si:dkey-151g10.6 | -0.032 |
| calm2b | 0.058 | si:dkey-51e6.1 | -0.031 |
| cirbpa | 0.057 | casq1a | -0.031 |
| csnk1a1 | 0.057 | si:ch73-367p23.2 | -0.031 |
| ptges3b | 0.057 | myoz1a | -0.031 |