"cathepsin S, ortholog 2, tandem duplicate 2"
ZFIN 
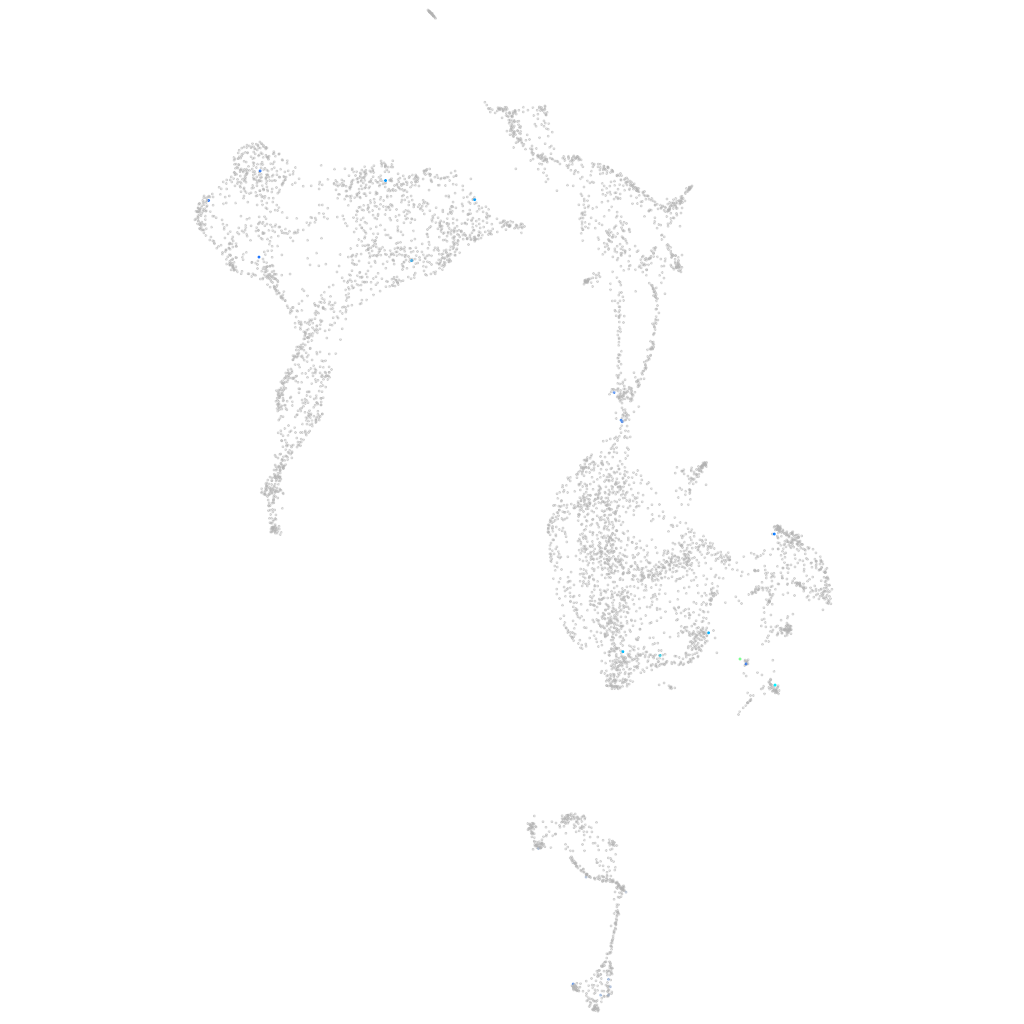
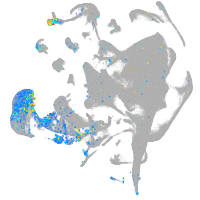
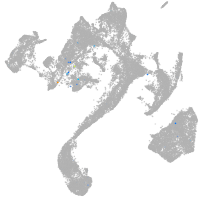
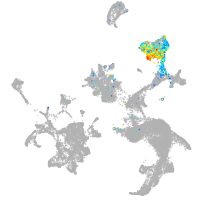
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| LOC110439308 | 0.435 | cfl1 | -0.051 |
| zgc:103700 | 0.429 | ssr3 | -0.041 |
| CU571315.2 | 0.406 | ptmab | -0.038 |
| havcr1 | 0.389 | arid3b | -0.037 |
| ccl34a.4 | 0.312 | ran | -0.037 |
| calhm1 | 0.307 | yeats4 | -0.035 |
| lpxn | 0.302 | psma2 | -0.035 |
| rhcga | 0.293 | ostc | -0.034 |
| BX908782.2 | 0.283 | aldoaa | -0.034 |
| LOC100535869 | 0.278 | sept15 | -0.033 |
| si:dkey-148a17.6 | 0.268 | prps1a | -0.032 |
| selp | 0.267 | mpc1 | -0.032 |
| ncf1 | 0.261 | snd1 | -0.032 |
| csgalnact1b | 0.257 | psma8 | -0.032 |
| spi1a | 0.249 | tmed10 | -0.031 |
| soat1 | 0.245 | tspan7 | -0.031 |
| galnt14 | 0.238 | ssr2 | -0.031 |
| CR788316.1 | 0.237 | si:dkey-250d21.1 | -0.030 |
| CU499330.1 | 0.235 | atp5f1c | -0.030 |
| cbln13 | 0.234 | srprb | -0.030 |
| arhgap27 | 0.234 | tmco1 | -0.030 |
| lygl1 | 0.234 | si:ch73-46j18.5 | -0.030 |
| CU929199.1 | 0.229 | cope | -0.030 |
| itprid1 | 0.225 | arid4a | -0.030 |
| coch | 0.224 | bzw1a | -0.030 |
| CABZ01068367.1 | 0.215 | fkbp11 | -0.030 |
| zgc:111983 | 0.215 | fkbp1aa | -0.029 |
| serpina1 | 0.214 | plod3 | -0.029 |
| si:dkey-5n18.1 | 0.214 | sar1b | -0.029 |
| cd74b | 0.211 | calr3b | -0.029 |
| cx28.9 | 0.211 | crtap | -0.029 |
| cldnj | 0.208 | fat1a | -0.029 |
| si:ch211-216b21.2 | 0.206 | h2afvb | -0.028 |
| zgc:92590 | 0.205 | ssr4 | -0.028 |
| eps8l1a | 0.201 | tmem39a | -0.028 |