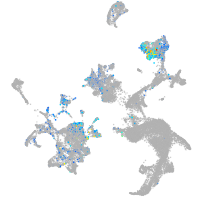
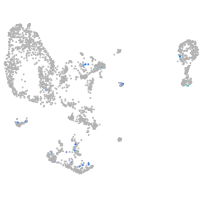
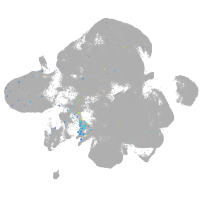
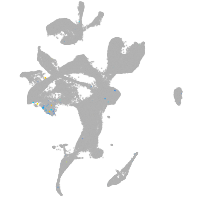
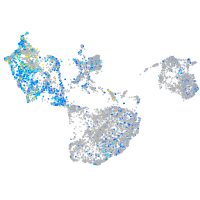
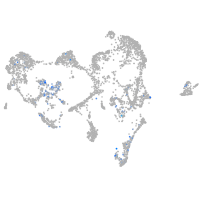
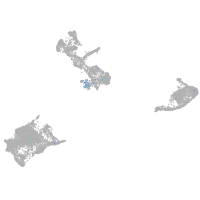
cathepsin K
ZFIN 
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| XLOC-040517 | 0.135 | cald1b | -0.106 |
| dcn | 0.132 | tpm2 | -0.097 |
| nphs1 | 0.132 | csrp1b | -0.097 |
| si:ch73-380l3.4 | 0.131 | desmb | -0.096 |
| sparc | 0.128 | cnn1b | -0.091 |
| col6a4a | 0.128 | mylkb | -0.090 |
| krt18b | 0.123 | acta2 | -0.088 |
| twist1b | 0.120 | tagln | -0.081 |
| LO017850.1 | 0.118 | gucy1a1 | -0.075 |
| foxl1 | 0.114 | ckbb | -0.075 |
| mfap5 | 0.113 | lmod1b | -0.073 |
| si:ch211-66i8.3 | 0.113 | acta1b | -0.073 |
| sema3bl | 0.112 | BX088707.3 | -0.073 |
| LOC108182814 | 0.112 | gapdhs | -0.071 |
| nr6a1b | 0.111 | myl6 | -0.071 |
| rasgrp2l | 0.111 | acta1a | -0.071 |
| col2a1b | 0.110 | pdlim3a | -0.070 |
| si:ch73-7i4.1 | 0.108 | XLOC-041870 | -0.069 |
| ctsla | 0.108 | tuba8l2 | -0.066 |
| lum | 0.108 | fhl1a | -0.066 |
| LOC101885558 | 0.107 | fbxl22 | -0.066 |
| glipr1a | 0.107 | itpka | -0.065 |
| vwde | 0.106 | hlx1 | -0.064 |
| mrap2a | 0.106 | pgm5 | -0.064 |
| col1a2 | 0.106 | myl9a | -0.063 |
| efnb1 | 0.105 | tpm1 | -0.063 |
| ndnfl | 0.104 | rgs2 | -0.062 |
| arsia | 0.104 | smtnb | -0.062 |
| alx4a | 0.103 | smtna | -0.061 |
| bace2 | 0.103 | zgc:110699 | -0.061 |
| col1a1a | 0.102 | vcla | -0.060 |
| bambib | 0.100 | si:ch211-137i24.10 | -0.060 |
| akap12b | 0.100 | pgam1a | -0.059 |
| si:dkeyp-2e4.5 | 0.100 | ak1 | -0.059 |
| grxcr1a.1 | 0.100 | sorbs3 | -0.058 |