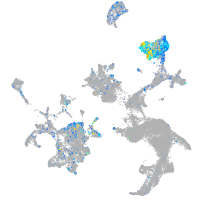
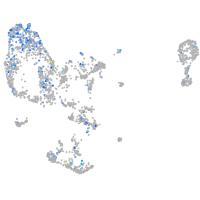
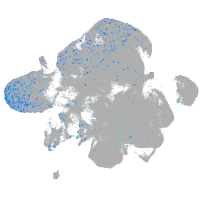
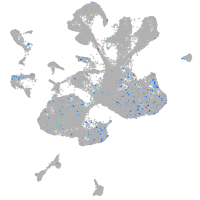
cathepsin C
ZFIN 
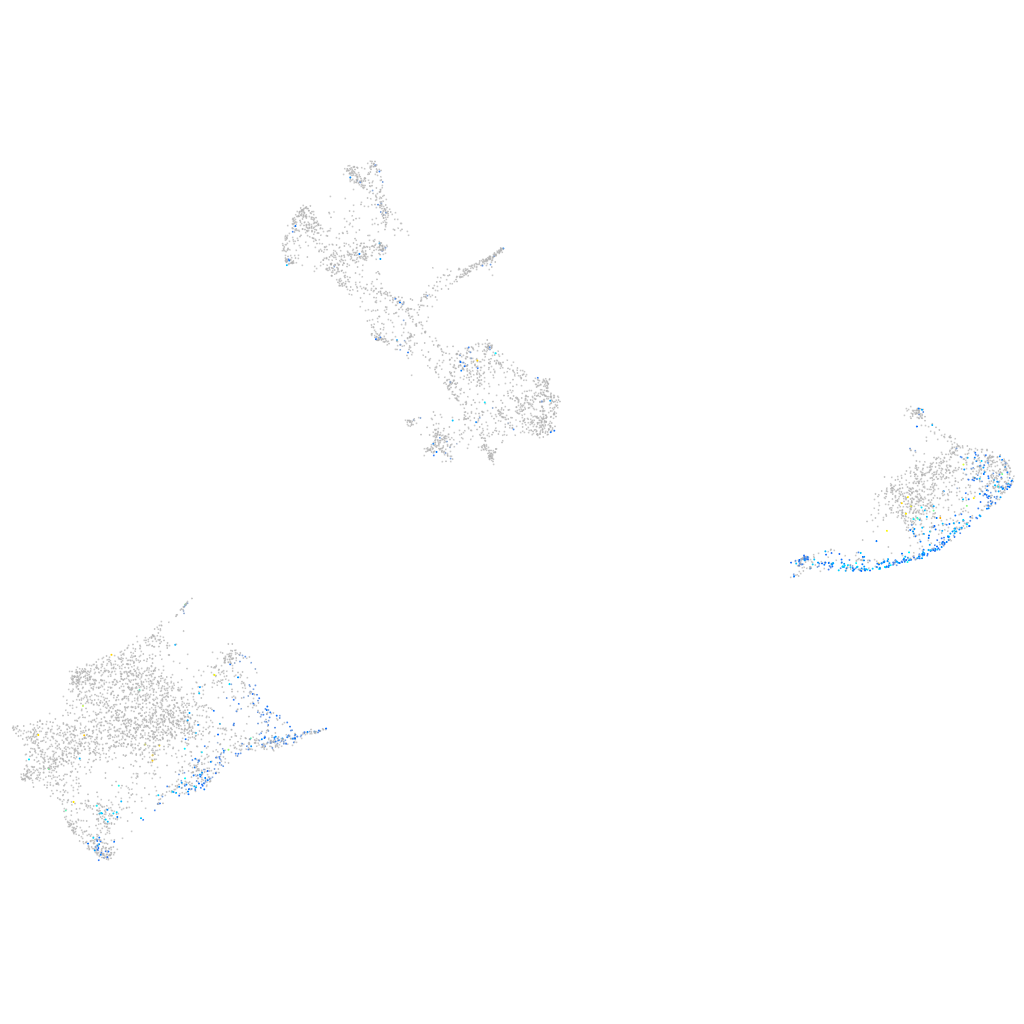
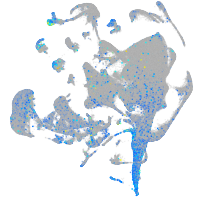
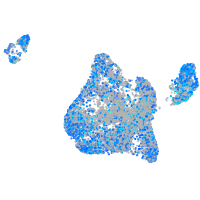
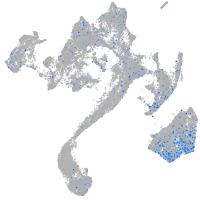
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| SPAG9 | 0.272 | pvalb2 | -0.123 |
| tmem243b | 0.268 | uraha | -0.122 |
| tyr | 0.265 | pvalb1 | -0.120 |
| lamp1a | 0.264 | mdkb | -0.106 |
| mitfa | 0.262 | defbl1 | -0.105 |
| slc24a5 | 0.261 | apoda.1 | -0.103 |
| slc3a2a | 0.251 | actc1b | -0.101 |
| msx1b | 0.250 | mdh1aa | -0.100 |
| slc7a5 | 0.249 | CABZ01021592.1 | -0.098 |
| tfap2e | 0.247 | hbae3 | -0.097 |
| tspan36 | 0.247 | gpnmb | -0.095 |
| dct | 0.243 | si:ch211-256m1.8 | -0.094 |
| slc37a2 | 0.242 | lypc | -0.092 |
| tyrp1a | 0.236 | krt18b | -0.091 |
| rabl6b | 0.236 | si:dkey-197i20.6 | -0.089 |
| phlda1 | 0.236 | paics | -0.088 |
| crestin | 0.235 | unm-sa821 | -0.087 |
| pah | 0.234 | stmn1b | -0.087 |
| atp6v0a2b | 0.233 | ptmaa | -0.085 |
| kita | 0.233 | si:dkey-251i10.2 | -0.085 |
| si:ch211-122f10.4 | 0.231 | hbbe1.3 | -0.084 |
| qdpra | 0.229 | mylpfa | -0.084 |
| slc45a2 | 0.229 | phyhd1 | -0.084 |
| sypl2b | 0.228 | gpm6aa | -0.084 |
| si:zfos-943e10.1 | 0.228 | hbae1.1 | -0.082 |
| degs1 | 0.227 | alx4a | -0.082 |
| glb1l | 0.226 | hbbe1.1 | -0.081 |
| pmela | 0.226 | tuba1c | -0.081 |
| tmem243a | 0.224 | akap12a | -0.079 |
| scpep1 | 0.223 | ponzr1 | -0.079 |
| klf6a | 0.223 | s100v2 | -0.078 |
| tspan10 | 0.222 | sytl2b | -0.078 |
| naga | 0.219 | alx4b | -0.077 |
| oca2 | 0.218 | gng3 | -0.076 |
| bace2 | 0.217 | si:ch73-89b15.3 | -0.076 |