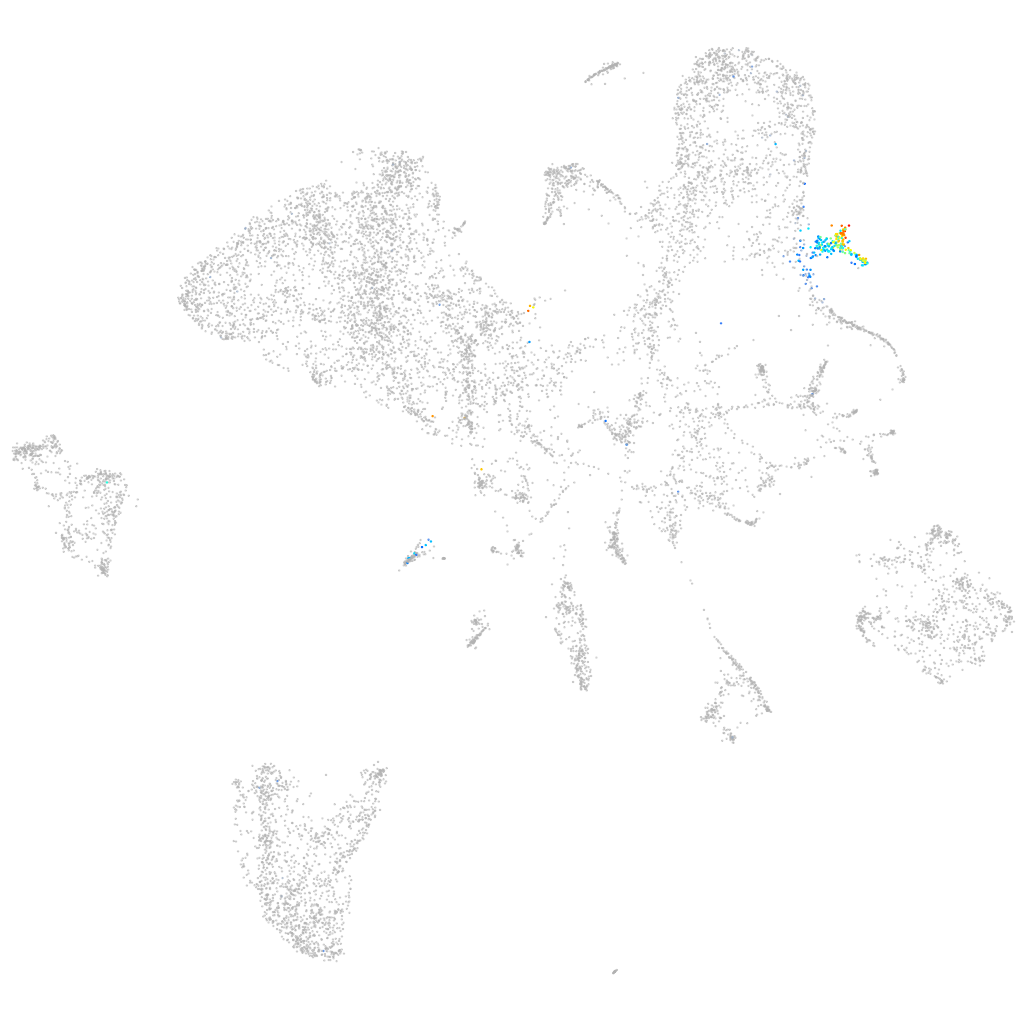
cathepsin Bb
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| LOC100537272 | 0.898 | bhmt | -0.121 |
| lrp2b | 0.884 | mat1a | -0.112 |
| cubn | 0.873 | aqp12 | -0.110 |
| ctsl.1 | 0.783 | si:dkey-16p21.8 | -0.108 |
| amn | 0.735 | apoc2 | -0.103 |
| dab2 | 0.725 | apoc1 | -0.101 |
| xirp1 | 0.722 | gatm | -0.099 |
| sptbn5 | 0.720 | apoa1b | -0.099 |
| slc6a18 | 0.700 | grhprb | -0.098 |
| lgmn | 0.688 | agxtb | -0.097 |
| tfeb | 0.669 | ybx1 | -0.097 |
| naga | 0.634 | kng1 | -0.095 |
| scpep1 | 0.617 | cdo1 | -0.094 |
| epdl2 | 0.597 | tfa | -0.093 |
| ms4a17a.17 | 0.592 | vtnb | -0.090 |
| snx2 | 0.585 | fabp3 | -0.090 |
| dpep1 | 0.575 | wu:fj16a03 | -0.090 |
| NAT16 (1 of many) | 0.563 | ndrg2 | -0.090 |
| ctsh | 0.555 | gamt | -0.089 |
| slc15a2 | 0.552 | serpinf2b | -0.089 |
| tspan34 | 0.547 | apoa2 | -0.089 |
| si:ch211-122f10.4 | 0.545 | ttc36 | -0.088 |
| CR381686.5 | 0.544 | serpina1 | -0.088 |
| rab32b | 0.541 | apobb.1 | -0.087 |
| fabp6 | 0.541 | apom | -0.087 |
| gnsb | 0.532 | cfhl4 | -0.087 |
| cpvl | 0.531 | zgc:123103 | -0.087 |
| si:dkey-28n18.9 | 0.524 | serpina1l | -0.087 |
| tpte | 0.510 | rbp2b | -0.087 |
| snx8a | 0.510 | mgst1.2 | -0.087 |
| si:dkey-194e6.1 | 0.505 | fgg | -0.086 |
| tmigd1 | 0.500 | hpda | -0.086 |
| zgc:174356 | 0.500 | fgb | -0.085 |
| grn1 | 0.477 | ambp | -0.085 |
| CU499336.2 | 0.476 | cnbpa | -0.085 |