cathepsin A
ZFIN 
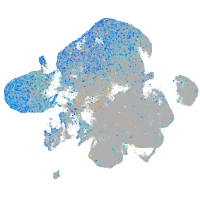
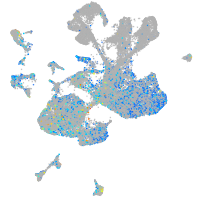
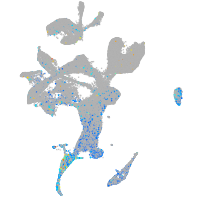
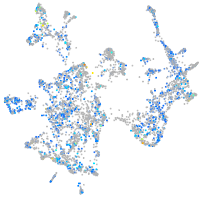
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| tspan36 | 0.337 | ptmab | -0.182 |
| slc45a2 | 0.324 | ptmaa | -0.159 |
| dct | 0.310 | ckbb | -0.156 |
| gpr143 | 0.309 | gpm6aa | -0.146 |
| pmela | 0.297 | tuba1c | -0.138 |
| tyrp1b | 0.282 | si:ch73-1a9.3 | -0.134 |
| rnaseka | 0.278 | hmgn6 | -0.127 |
| bace2 | 0.273 | snap25b | -0.124 |
| agtrap | 0.271 | epb41a | -0.121 |
| tmem88b | 0.271 | gapdhs | -0.115 |
| zgc:110591 | 0.271 | syt5b | -0.114 |
| cx43 | 0.271 | atp6v0cb | -0.110 |
| tfec | 0.271 | rnasekb | -0.110 |
| tyrp1a | 0.269 | ndrg4 | -0.109 |
| qdpra | 0.267 | foxg1b | -0.108 |
| pmelb | 0.265 | gpm6ab | -0.105 |
| tspan10 | 0.261 | stmn1b | -0.104 |
| pcbd1 | 0.260 | sypb | -0.104 |
| trpm1b | 0.256 | crx | -0.102 |
| pttg1ipb | 0.254 | hmgb1a | -0.101 |
| rab38 | 0.253 | cadm3 | -0.100 |
| oca2 | 0.252 | neurod4 | -0.100 |
| mitfa | 0.251 | atp1b2a | -0.098 |
| gstp1 | 0.251 | rtn1a | -0.098 |
| tyr | 0.250 | mpp6b | -0.098 |
| cd63 | 0.249 | pvalb1 | -0.096 |
| wls | 0.248 | nova2 | -0.094 |
| zgc:110239 | 0.245 | vamp2 | -0.093 |
| sparc | 0.243 | vsx1 | -0.093 |
| slc24a5 | 0.241 | scrt2 | -0.092 |
| rab32a | 0.238 | calb2b | -0.092 |
| FP085398.1 | 0.238 | rs1a | -0.091 |
| cracr2ab | 0.238 | pvalb2 | -0.091 |
| fabp11a | 0.228 | atpv0e2 | -0.090 |
| sytl2b | 0.227 | h3f3c | -0.089 |