"CTR9 homolog, Paf1/RNA polymerase II complex component"
ZFIN 
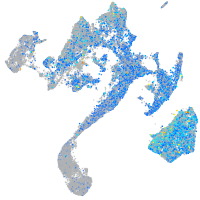
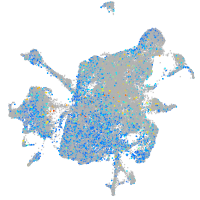
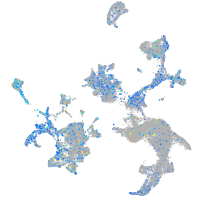
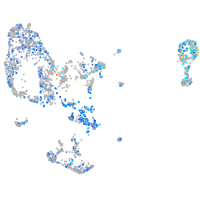
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| hspb1 | 0.285 | si:ch211-139a5.9 | -0.237 |
| wu:fb97g03 | 0.284 | ldhba | -0.214 |
| apoeb | 0.267 | rpl37 | -0.214 |
| apoc1 | 0.267 | atp1b1a | -0.203 |
| seta | 0.267 | eno3 | -0.200 |
| setb | 0.262 | atp5mc3b | -0.196 |
| hmga1a | 0.261 | ahcy | -0.194 |
| hmgb2b | 0.261 | atp5fa1 | -0.193 |
| eif2s1a | 0.260 | mdh1aa | -0.193 |
| snrpe | 0.258 | rps10 | -0.192 |
| cdx4 | 0.255 | atp5f1b | -0.192 |
| si:ch73-281n10.2 | 0.250 | si:dkey-16p21.8 | -0.188 |
| anp32b | 0.249 | aldob | -0.186 |
| ppig | 0.249 | aldh6a1 | -0.184 |
| nucks1a | 0.248 | atp5mc1 | -0.182 |
| hmgb2a | 0.247 | tpi1b | -0.179 |
| hnrnpabb | 0.247 | zgc:114188 | -0.178 |
| znfl2a | 0.247 | gapdh | -0.175 |
| khdrbs1a | 0.242 | atp5l | -0.173 |
| syncrip | 0.242 | atp1a1a.4 | -0.170 |
| ddx39ab | 0.242 | suclg1 | -0.169 |
| hnrnpaba | 0.239 | ndufa4l | -0.169 |
| eif5a2 | 0.238 | cdaa | -0.168 |
| ebna1bp2 | 0.237 | atp5f1e | -0.168 |
| smc1al | 0.237 | rps17 | -0.167 |
| qkia | 0.235 | romo1 | -0.166 |
| fbl | 0.234 | COX3 | -0.165 |
| ncl | 0.234 | cox6c | -0.164 |
| si:ch73-1a9.3 | 0.234 | zgc:158463 | -0.163 |
| ptmab | 0.232 | cdh17 | -0.163 |
| snrpd1 | 0.231 | grhprb | -0.162 |
| rbm8a | 0.230 | atp5if1b | -0.162 |
| nop56 | 0.229 | cox6b1 | -0.162 |
| srsf1a | 0.229 | ndufb3 | -0.161 |
| hmgn6 | 0.228 | mt-atp6 | -0.160 |