"CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase like 3"
ZFIN 
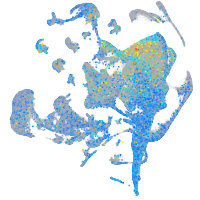
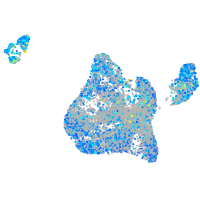
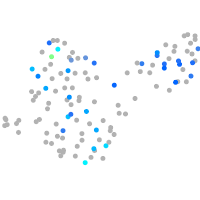
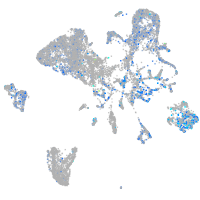
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| si:ch211-69g19.2 | 0.191 | slc1a2b | -0.099 |
| ccnb1 | 0.189 | fabp7a | -0.082 |
| cdc20 | 0.189 | atp1a1b | -0.081 |
| kpna2 | 0.186 | ckbb | -0.078 |
| plk1 | 0.184 | ptn | -0.070 |
| tpx2 | 0.183 | mdkb | -0.066 |
| ube2c | 0.182 | ppap2d | -0.066 |
| mki67 | 0.182 | slc1a3b | -0.066 |
| cenpf | 0.181 | CU467822.1 | -0.064 |
| nusap1 | 0.180 | si:ch211-66e2.5 | -0.062 |
| aspm | 0.179 | gpr37l1b | -0.062 |
| kif11 | 0.179 | cx43 | -0.062 |
| aurka | 0.179 | slc3a2a | -0.060 |
| kifc1 | 0.177 | glula | -0.059 |
| tacc3 | 0.175 | gapdhs | -0.059 |
| prc1a | 0.175 | slc4a4a | -0.058 |
| mad2l1 | 0.175 | hepacama | -0.057 |
| cdk1 | 0.175 | slc6a1b | -0.057 |
| aurkb | 0.174 | atp1b4 | -0.056 |
| kmt5ab | 0.171 | nfia | -0.056 |
| ccnb3 | 0.170 | COX3 | -0.055 |
| top2a | 0.169 | pvalb2 | -0.055 |
| g2e3 | 0.167 | sept8b | -0.054 |
| cdca8 | 0.165 | pvalb1 | -0.054 |
| ccna2 | 0.161 | efhd1 | -0.053 |
| armc1l | 0.159 | slc6a11b | -0.053 |
| pif1 | 0.156 | cspg5b | -0.053 |
| birc5a | 0.155 | cdo1 | -0.053 |
| spdl1 | 0.155 | cspg5a | -0.051 |
| kif23 | 0.154 | acbd7 | -0.051 |
| cenpe | 0.152 | CR848047.1 | -0.051 |
| kif20a | 0.150 | qki2 | -0.051 |
| dlgap5 | 0.149 | ndrg3a | -0.051 |
| hmmr | 0.149 | anxa13 | -0.051 |
| kif22 | 0.145 | NPAS3 | -0.050 |