cysteine-rich with EGF-like domains 1b
ZFIN 
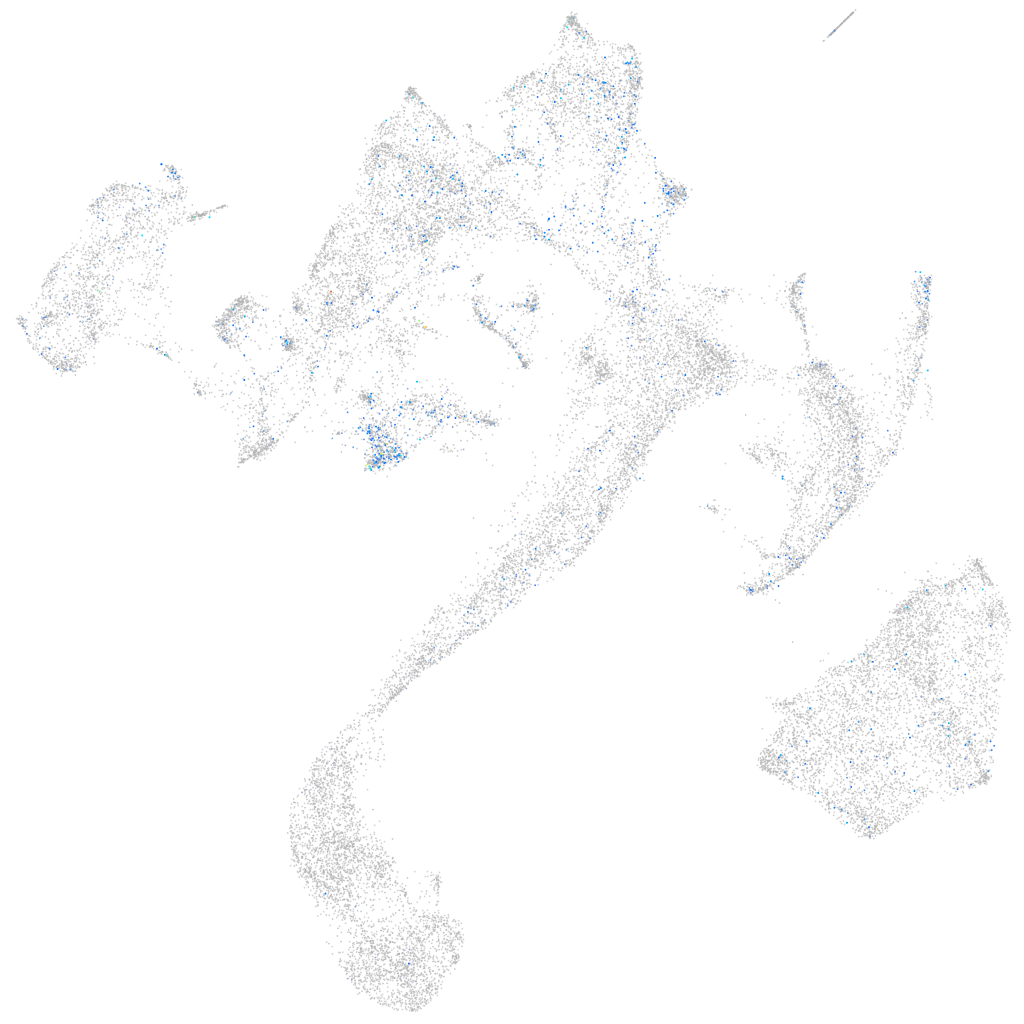
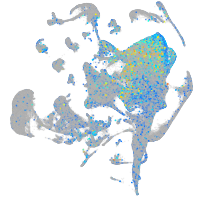
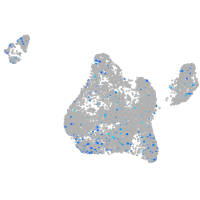
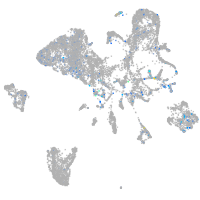
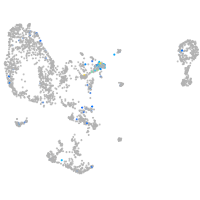
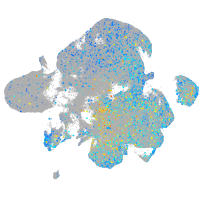
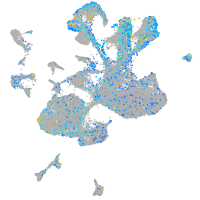
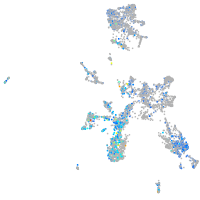
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| rtn1b | 0.149 | myl1 | -0.053 |
| gng3 | 0.144 | myoz1b | -0.053 |
| zgc:65894 | 0.143 | mylpfb | -0.052 |
| rnasekb | 0.138 | myom2a | -0.051 |
| gpm6aa | 0.135 | CABZ01061524.1 | -0.051 |
| stmn1b | 0.132 | casq1a | -0.050 |
| tubb5 | 0.132 | tnnt3a | -0.049 |
| stxbp1a | 0.131 | tnni2a.4 | -0.049 |
| ptmaa | 0.129 | slc25a4 | -0.049 |
| fez1 | 0.128 | ryr3 | -0.049 |
| LOC101887017 | 0.128 | si:ch73-367p23.2 | -0.048 |
| sncb | 0.127 | hspb1 | -0.048 |
| vamp2 | 0.127 | ttn.1 | -0.048 |
| tuba1c | 0.126 | zgc:101853 | -0.047 |
| elavl3 | 0.124 | mylz3 | -0.047 |
| calr | 0.124 | hhatla | -0.047 |
| stmn2a | 0.124 | tnnc2 | -0.047 |
| olfm1b | 0.122 | mybphb | -0.047 |
| cspg5a | 0.122 | gamt | -0.046 |
| stx1b | 0.122 | tnnt3b | -0.046 |
| maptb | 0.122 | tpma | -0.045 |
| gap43 | 0.120 | ckmb | -0.045 |
| carmil2 | 0.117 | si:ch211-255p10.3 | -0.045 |
| celf4 | 0.116 | ldb3b | -0.045 |
| si:ch211-214j24.9 | 0.115 | prr33 | -0.044 |
| elavl4 | 0.112 | eef2l2 | -0.044 |
| rundc3ab | 0.111 | XLOC-025819 | -0.044 |
| zgc:153426 | 0.111 | myoz2a | -0.043 |
| cnrip1a | 0.111 | ckma | -0.043 |
| atp1a3a | 0.110 | myhz1.3 | -0.043 |
| jagn1a | 0.110 | pvalb2 | -0.043 |
| evlb | 0.109 | igfn1.1 | -0.043 |
| gapdhs | 0.109 | rpl6 | -0.043 |
| hspa5 | 0.108 | dhrs7cb | -0.042 |
| id4 | 0.108 | XLOC-001975 | -0.042 |