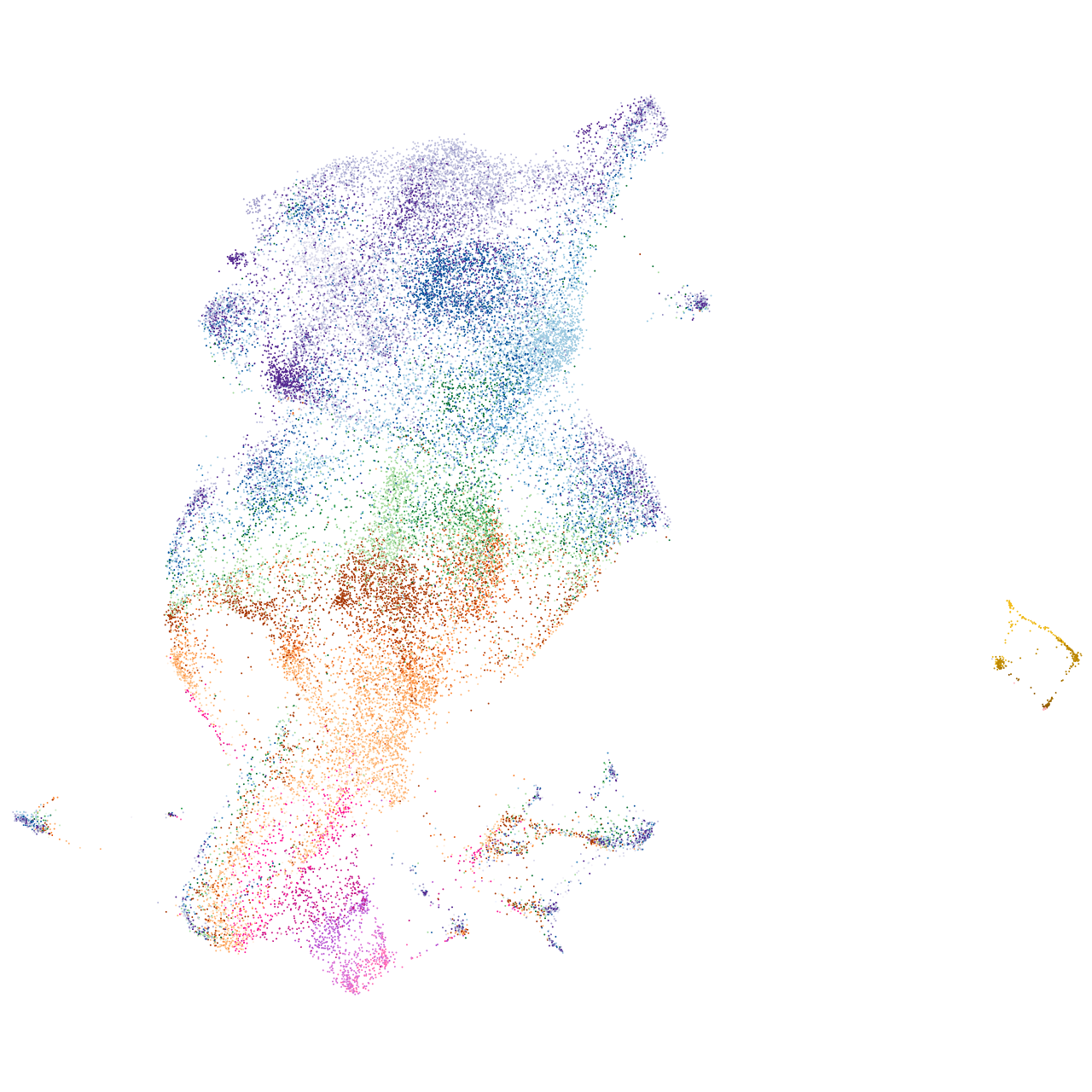CREB binding protein b
ZFIN 
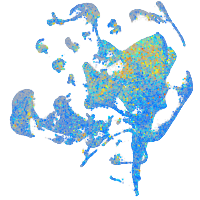
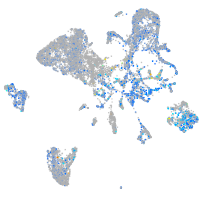
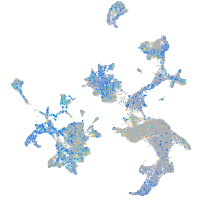
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| NC-002333.4 | 0.128 | cyt1 | -0.106 |
| CR383676.1 | 0.119 | si:ch211-195b11.3 | -0.091 |
| mt-cyb | 0.113 | icn2 | -0.088 |
| mt-co1 | 0.109 | krt4 | -0.087 |
| mt-co2 | 0.109 | cyt1l | -0.074 |
| COX3 | 0.103 | rpl37 | -0.071 |
| nucks1a | 0.101 | rpl7 | -0.070 |
| acin1a | 0.099 | krtt1c19e | -0.069 |
| mt-atp6 | 0.099 | icn | -0.068 |
| cdh1 | 0.098 | rpl39 | -0.068 |
| mt-nd1 | 0.095 | btf3 | -0.068 |
| pnrc2 | 0.094 | krt17 | -0.067 |
| marcksl1b | 0.093 | rps10 | -0.066 |
| syncrip | 0.092 | faua | -0.066 |
| f11r.1 | 0.091 | ahcy | -0.065 |
| khdrbs1a | 0.091 | rpl21 | -0.064 |
| si:ch211-288g17.3 | 0.090 | anxa1a | -0.062 |
| evpla | 0.090 | rps24 | -0.061 |
| akap12b | 0.089 | rps2 | -0.061 |
| ankrd11 | 0.088 | rpl35 | -0.059 |
| mt-nd4 | 0.088 | rpl8 | -0.059 |
| hmga1a | 0.088 | romo1 | -0.058 |
| mt-nd2 | 0.087 | rpl22 | -0.058 |
| mki67 | 0.087 | rplp1 | -0.058 |
| hnrnpa1a | 0.087 | rpl36a | -0.057 |
| itsn2b | 0.087 | zgc:114188 | -0.057 |
| smarca4a | 0.086 | rpl10 | -0.056 |
| marcksb | 0.086 | rps23 | -0.055 |
| pleca | 0.085 | rplp0 | -0.054 |
| chd4b | 0.084 | rpl31 | -0.054 |
| epcam | 0.084 | rps26l | -0.054 |
| hnrnpa0b | 0.082 | rpl17 | -0.053 |
| ctnnd1 | 0.082 | rpl10a | -0.053 |
| chd7 | 0.081 | s100a10b | -0.053 |
| glulb | 0.081 | rps5 | -0.052 |