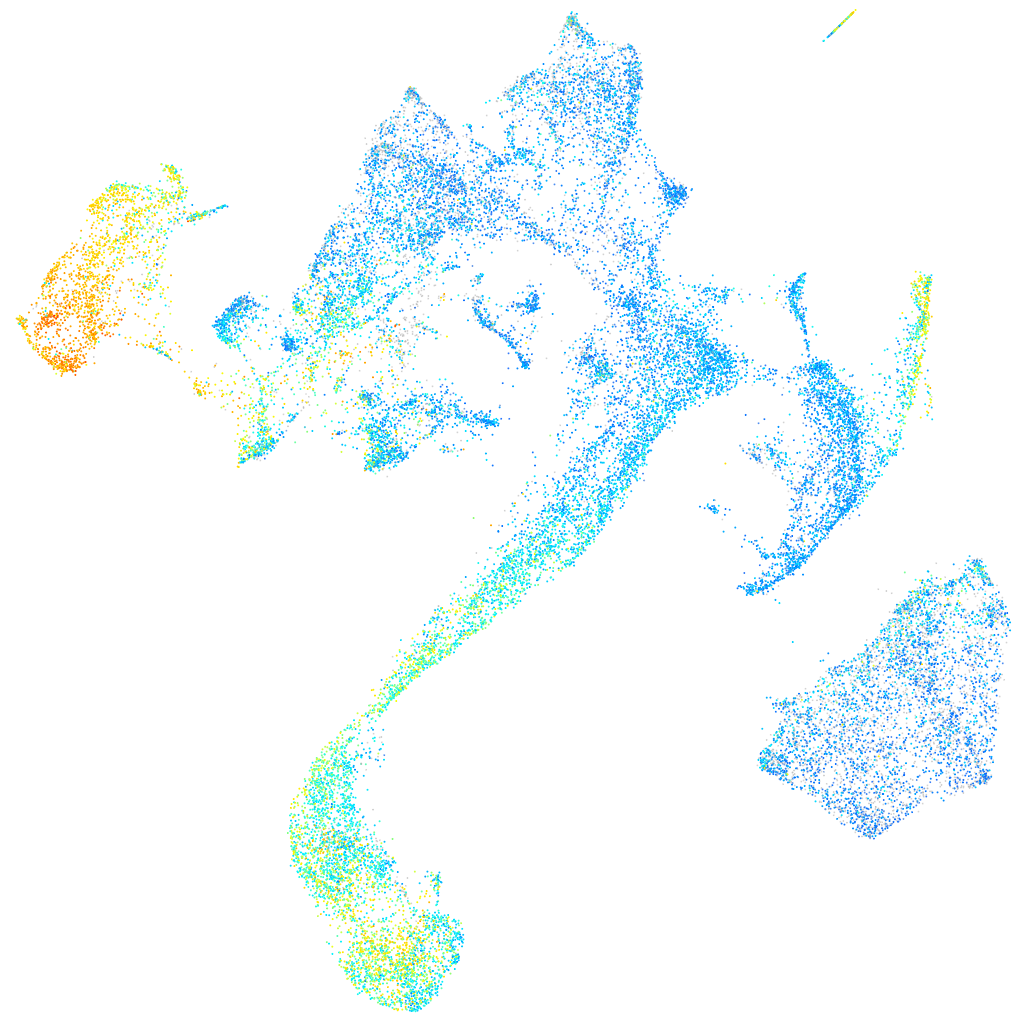cytochrome c oxidase subunit 7A2a
ZFIN 
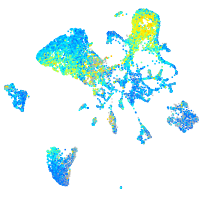
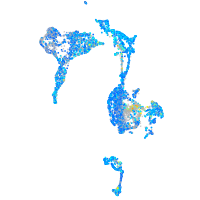
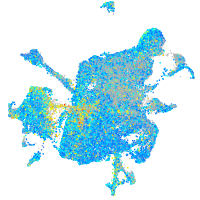
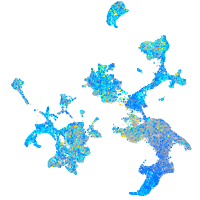
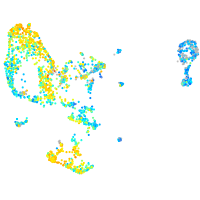
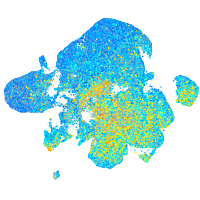
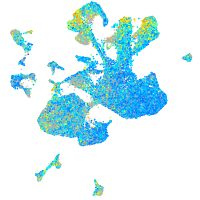
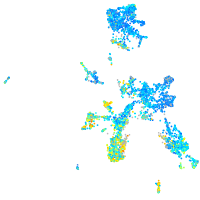
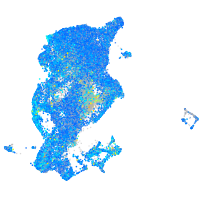
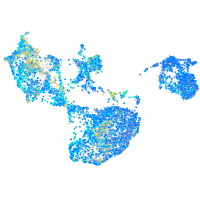
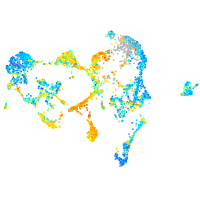
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| aldoab | 0.634 | si:ch73-1a9.3 | -0.553 |
| atp5mc1 | 0.611 | hsp90ab1 | -0.551 |
| atp5meb | 0.609 | ptmab | -0.536 |
| actc1b | 0.605 | pabpc1a | -0.519 |
| atp5if1b | 0.604 | si:ch211-222l21.1 | -0.501 |
| atp5f1b | 0.602 | cirbpb | -0.477 |
| ttn.2 | 0.598 | h3f3d | -0.472 |
| ttn.1 | 0.597 | fthl27 | -0.468 |
| ak1 | 0.595 | hmgb2b | -0.465 |
| tmem38a | 0.594 | hmgb2a | -0.456 |
| atp2a1 | 0.588 | khdrbs1a | -0.448 |
| ckmb | 0.584 | anp32a | -0.447 |
| gapdh | 0.583 | cirbpa | -0.445 |
| atp5mc3b | 0.583 | si:ch73-281n10.2 | -0.437 |
| srl | 0.583 | marcksl1b | -0.436 |
| suclg1 | 0.583 | hnrnpaba | -0.434 |
| ckma | 0.581 | actb2 | -0.429 |
| cox6a1 | 0.578 | ubc | -0.428 |
| cox5aa | 0.571 | hmga1a | -0.424 |
| eno3 | 0.568 | hmgn7 | -0.422 |
| desma | 0.565 | tuba8l4 | -0.422 |
| mdh2 | 0.565 | hnrnpa0b | -0.421 |
| COX5B | 0.565 | nucks1a | -0.421 |
| casq2 | 0.562 | hmgn2 | -0.419 |
| neb | 0.562 | h2afvb | -0.415 |
| atp5l | 0.561 | si:ch211-288g17.3 | -0.414 |
| acta1a | 0.561 | tmsb4x | -0.413 |
| pgam2 | 0.559 | seta | -0.413 |
| atp5pd | 0.552 | anp32b | -0.413 |
| cav3 | 0.551 | cx43.4 | -0.412 |
| atp5pf | 0.551 | acin1a | -0.405 |
| actn3b | 0.548 | ran | -0.402 |
| chchd10 | 0.547 | syncrip | -0.398 |
| nme2b.2 | 0.547 | hnrnpa1b | -0.389 |
| cox8a | 0.546 | hnrnpabb | -0.388 |