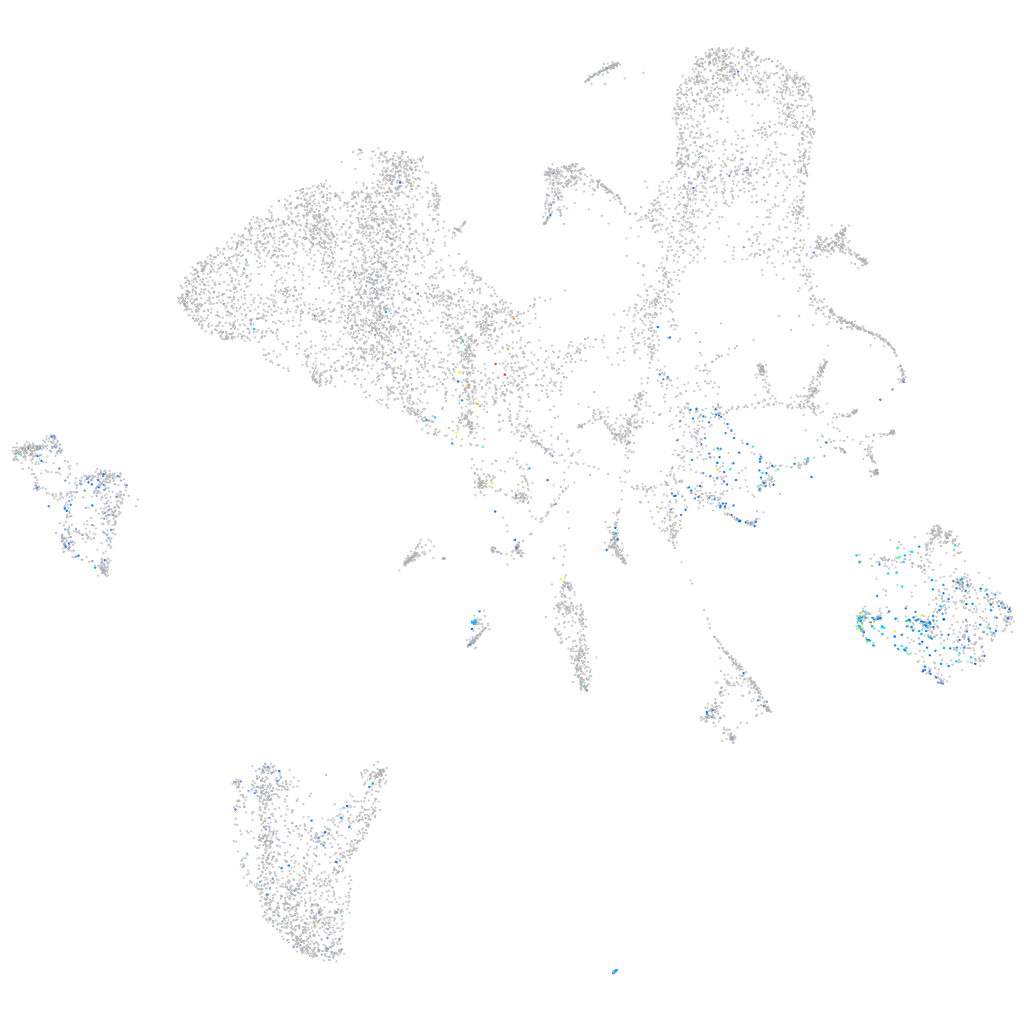
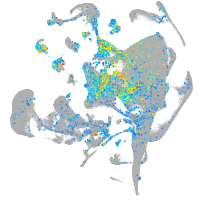
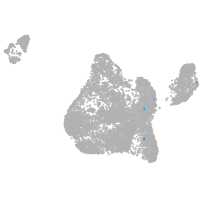
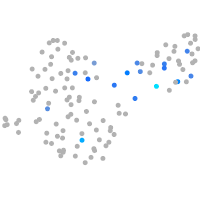
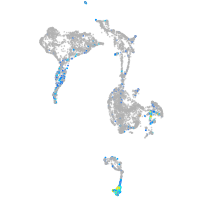
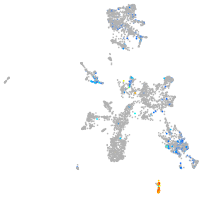
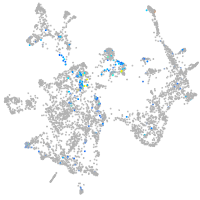
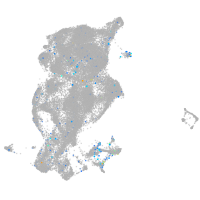
cytochrome c oxidase subunit 4I2
ZFIN 
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| pltp | 0.308 | rpl37 | -0.210 |
| si:ch211-152c2.3 | 0.283 | rps10 | -0.208 |
| akap12b | 0.278 | eno3 | -0.196 |
| cdh6 | 0.271 | zgc:114188 | -0.190 |
| cyp26c1 | 0.263 | ahcy | -0.185 |
| hspb1 | 0.262 | nme2b.1 | -0.178 |
| marcksl1b | 0.251 | gapdh | -0.176 |
| mcama | 0.240 | rps17 | -0.176 |
| si:dkey-43p13.5 | 0.236 | suclg1 | -0.172 |
| asph | 0.232 | gamt | -0.165 |
| marcksb | 0.230 | atp5if1b | -0.162 |
| apela | 0.227 | atp5mc1 | -0.162 |
| lima1a | 0.227 | mt-atp6 | -0.160 |
| nkx2.7 | 0.223 | prdx2 | -0.159 |
| qkia | 0.219 | sod1 | -0.157 |
| cxcl12b | 0.215 | tpi1b | -0.157 |
| ctnnb1 | 0.214 | gpx4a | -0.157 |
| nr6a1a | 0.213 | fbp1b | -0.156 |
| si:ch73-281n10.2 | 0.213 | pklr | -0.156 |
| irx7 | 0.212 | gstt1a | -0.155 |
| hmgn6 | 0.211 | cox6b1 | -0.150 |
| syncrip | 0.208 | apoa4b.1 | -0.150 |
| acin1a | 0.208 | atp5f1b | -0.150 |
| nucks1a | 0.207 | BX908782.3 | -0.149 |
| hnrnpub | 0.207 | atp5mc3b | -0.148 |
| fzd7a | 0.207 | lgals2b | -0.146 |
| prtfdc1 | 0.207 | apoc2 | -0.146 |
| hnrnpa1a | 0.206 | cox6a1 | -0.146 |
| NC-002333.4 | 0.206 | mt-nd1 | -0.145 |
| prdx5 | 0.205 | mdh1aa | -0.144 |
| ppig | 0.204 | glud1b | -0.142 |
| gpm6ab | 0.203 | scp2a | -0.142 |
| hmgb2a | 0.203 | atp5fa1 | -0.142 |
| anp32a | 0.202 | cx32.3 | -0.140 |
| sept15 | 0.201 | pgk1 | -0.139 |