"coronin, actin binding protein, 1Cb"
ZFIN 
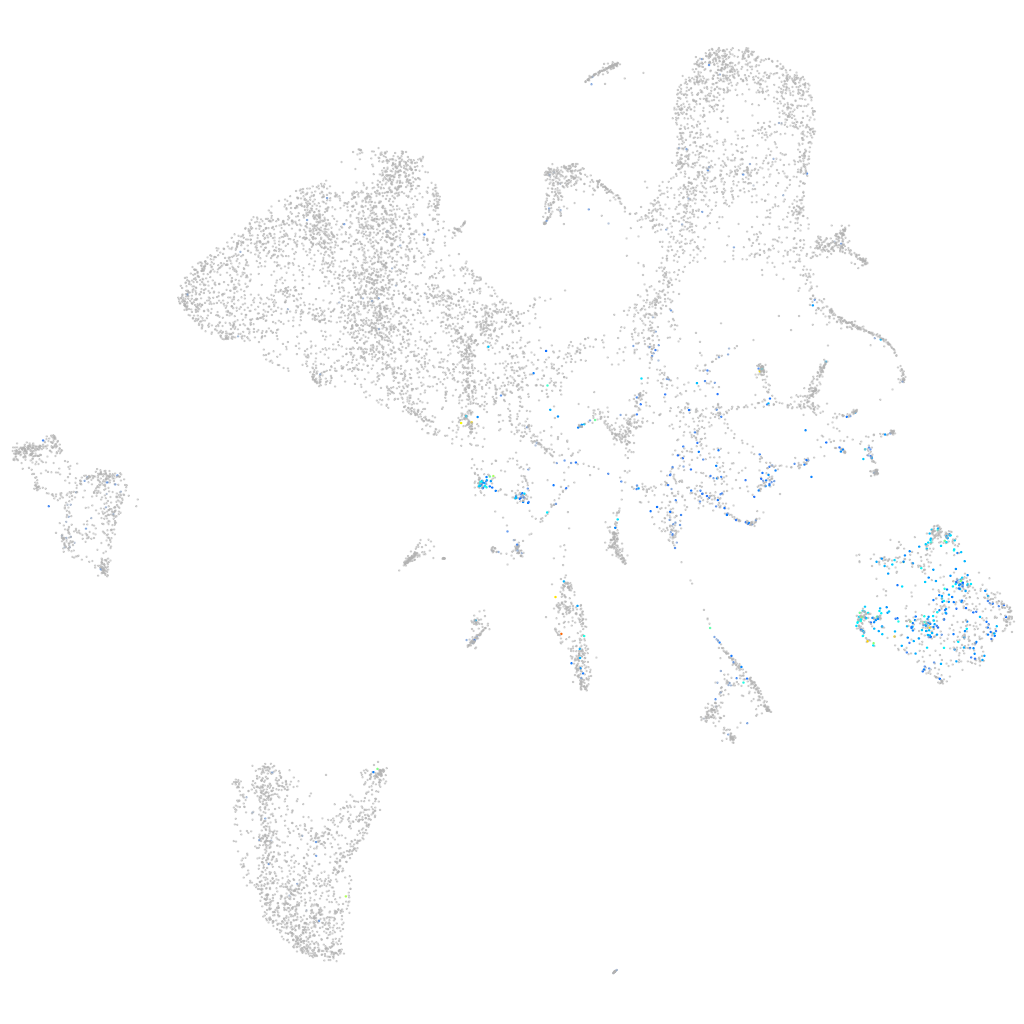
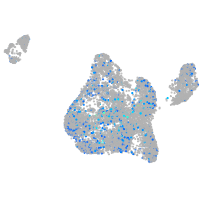
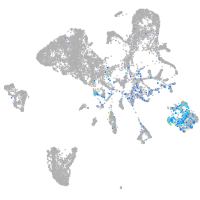
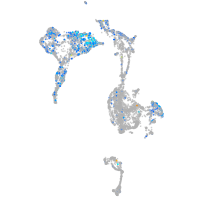
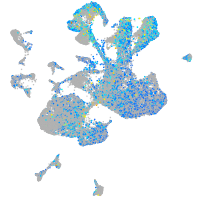
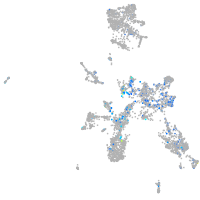
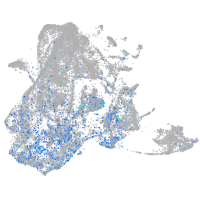
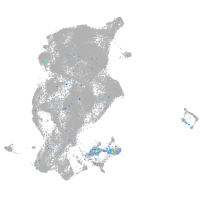
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| marcksb | 0.371 | rpl37 | -0.295 |
| marcksl1b | 0.346 | rps10 | -0.291 |
| hspb1 | 0.340 | zgc:114188 | -0.269 |
| akap12b | 0.324 | rps17 | -0.253 |
| si:ch211-152c2.3 | 0.323 | nme2b.1 | -0.244 |
| cx43.4 | 0.321 | ahcy | -0.220 |
| nucks1a | 0.316 | eno3 | -0.219 |
| asph | 0.310 | sod1 | -0.216 |
| hnrnpa1a | 0.307 | gapdh | -0.210 |
| hnrnpub | 0.304 | eef1da | -0.207 |
| ilf3b | 0.304 | atp5if1b | -0.199 |
| nr6a1a | 0.300 | gamt | -0.194 |
| hnrnpa1b | 0.299 | zgc:92744 | -0.192 |
| si:ch211-137a8.4 | 0.297 | suclg1 | -0.191 |
| NC-002333.4 | 0.296 | prdx2 | -0.184 |
| acin1a | 0.295 | atp5fa1 | -0.182 |
| hmgb1b | 0.295 | atp5mc3b | -0.182 |
| ppig | 0.294 | aldob | -0.178 |
| lima1a | 0.294 | gstp1 | -0.178 |
| seta | 0.293 | atp5mc1 | -0.175 |
| hmgn6 | 0.293 | tpi1b | -0.174 |
| COX7A2 | 0.292 | atp5f1b | -0.173 |
| cdh6 | 0.291 | nupr1b | -0.172 |
| syncrip | 0.291 | aldh6a1 | -0.171 |
| snrnp70 | 0.289 | glud1b | -0.171 |
| hmgb3a | 0.288 | fbp1b | -0.171 |
| pltp | 0.288 | atp5pf | -0.171 |
| tardbp | 0.287 | dap | -0.170 |
| hnrnpa0a | 0.286 | atp5l | -0.168 |
| anp32a | 0.284 | mdh1aa | -0.167 |
| top1l | 0.283 | pklr | -0.167 |
| hnrnpaba | 0.282 | COX7A2 (1 of many) | -0.167 |
| ewsr1a | 0.280 | gpx4a | -0.166 |
| sox11b | 0.276 | cox6a1 | -0.166 |
| vox | 0.276 | mt-atp6 | -0.165 |