coenzyme Q9 homolog (S. cerevisiae)
ZFIN 
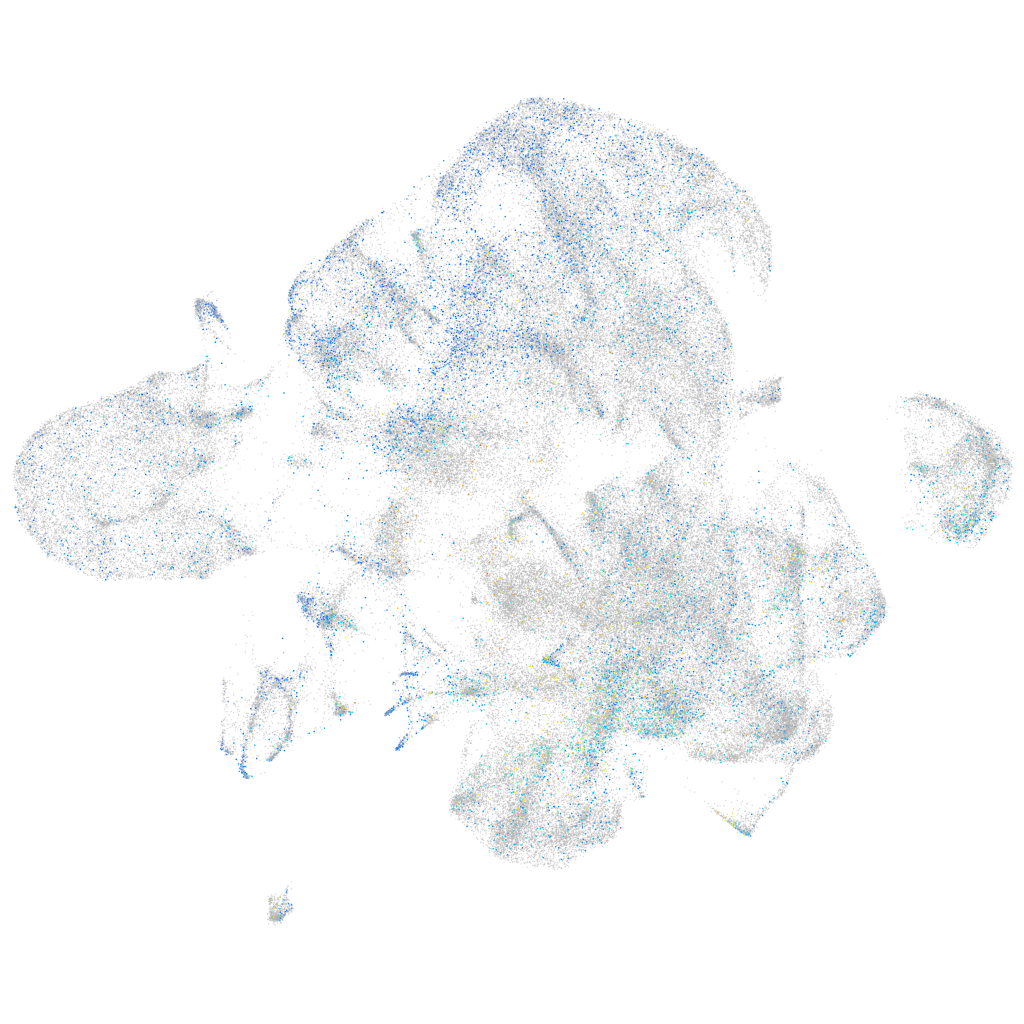
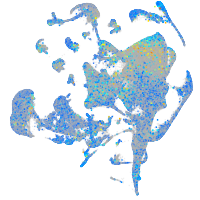
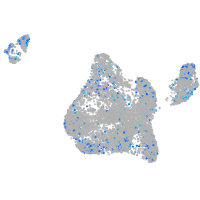
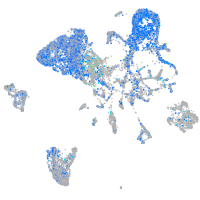
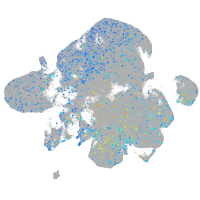
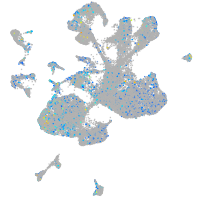
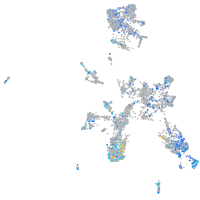
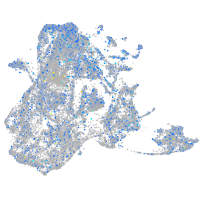
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| pkma | 0.080 | si:dkey-56m19.5 | -0.056 |
| tpi1b | 0.079 | si:ch211-222l21.1 | -0.056 |
| gapdhs | 0.079 | cirbpb | -0.050 |
| calm1b | 0.078 | sox11b | -0.048 |
| gpia | 0.076 | ptmab | -0.048 |
| eno1a | 0.075 | dlb | -0.047 |
| aldocb | 0.073 | hmga1a | -0.046 |
| sncgb | 0.073 | chd4a | -0.043 |
| pgk1 | 0.072 | FO082781.1 | -0.041 |
| ldhba | 0.071 | hmgn6 | -0.039 |
| ndufa4 | 0.070 | khdrbs1a | -0.038 |
| syngr1a | 0.068 | cdkn1ca | -0.038 |
| ndrg3a | 0.068 | elavl3 | -0.036 |
| calm3a | 0.068 | myt1a | -0.035 |
| atp5if1b | 0.067 | ebf2 | -0.035 |
| atp1b1b | 0.065 | rplp2l | -0.035 |
| sh3gl2a | 0.064 | rps12 | -0.035 |
| atp6v1b2 | 0.064 | rcc2 | -0.034 |
| chchd10 | 0.064 | sox11a | -0.033 |
| atp5meb | 0.064 | insm1a | -0.033 |
| sod2 | 0.063 | tp53inp2 | -0.033 |
| eno2 | 0.063 | si:ch73-1a9.3 | -0.033 |
| syn2a | 0.062 | dla | -0.033 |
| nptna | 0.062 | hnrnpa1a | -0.032 |
| si:ch73-119p20.1 | 0.062 | notch1a | -0.032 |
| smdt1a | 0.062 | hnrnpub | -0.031 |
| atp5mc1 | 0.062 | tmsb | -0.031 |
| atp5mc3b | 0.062 | NC-002333.4 | -0.031 |
| nsfa | 0.062 | gadd45gb.1 | -0.031 |
| COX5B | 0.061 | scrt2 | -0.031 |
| atp6v0d1 | 0.060 | peli1b | -0.031 |
| atp5f1b | 0.060 | rtca | -0.030 |
| atp5l | 0.059 | hnrnpm | -0.030 |
| atp2b3b | 0.058 | LOC798783 | -0.030 |
| ywhag2 | 0.058 | si:ch211-288g17.3 | -0.030 |