COMM domain containing 4
ZFIN 
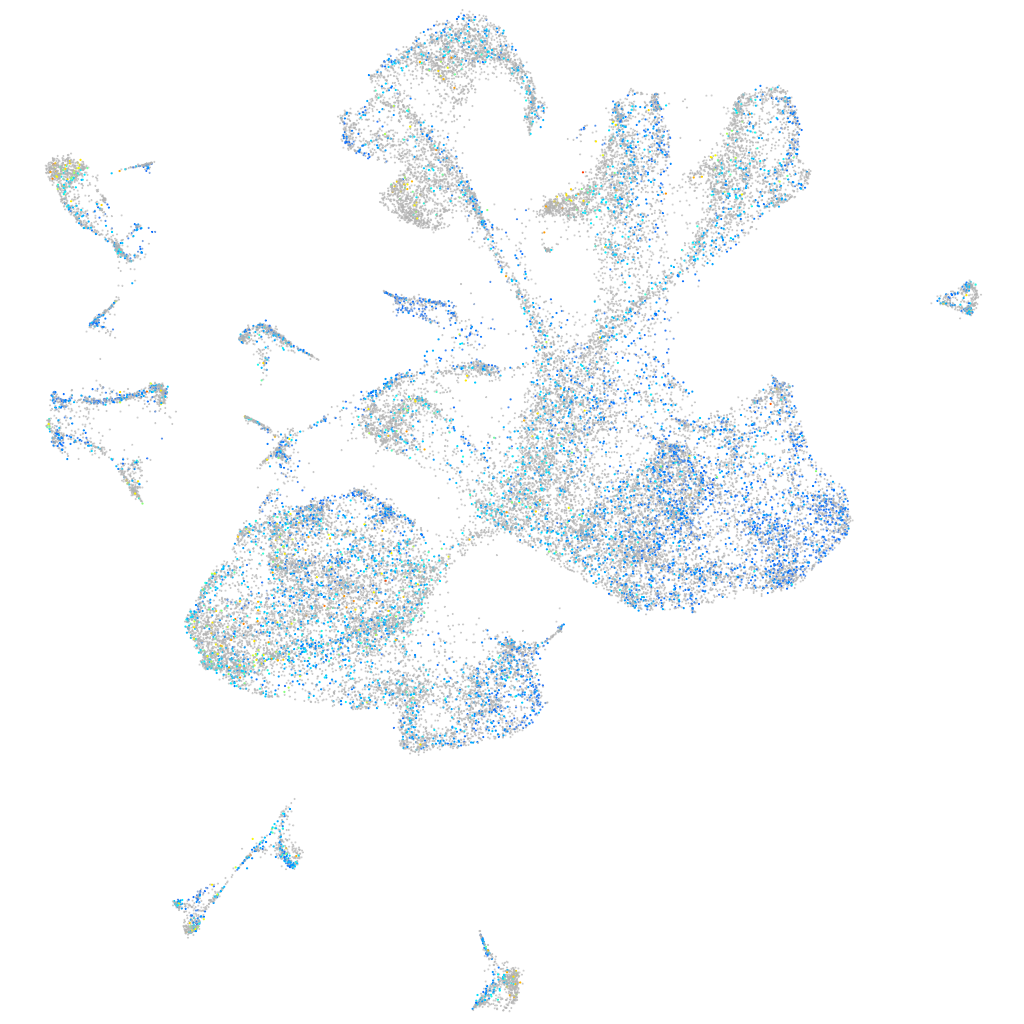
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| vamp3 | 0.074 | NC-002333.4 | -0.048 |
| zgc:153867 | 0.070 | elavl3 | -0.048 |
| atp5pd | 0.070 | onecut1 | -0.044 |
| dbi | 0.069 | isl2a | -0.042 |
| sept10 | 0.069 | pik3r3b | -0.041 |
| atp5mc3b | 0.068 | elavl4 | -0.041 |
| hsbp1b | 0.068 | myt1b | -0.039 |
| myl12.1 | 0.068 | COX3 | -0.038 |
| GCA | 0.068 | si:ch73-386h18.1 | -0.038 |
| ywhabb | 0.068 | islr2 | -0.037 |
| actb2 | 0.067 | nova2 | -0.037 |
| tubb4b | 0.067 | aplp1 | -0.037 |
| anxa13 | 0.067 | golga7ba | -0.036 |
| ssr3 | 0.066 | isl1 | -0.035 |
| sdcbp2 | 0.066 | LOC100537384 | -0.034 |
| atp5pb | 0.065 | FO082781.1 | -0.034 |
| arf2b | 0.065 | CABZ01072614.1 | -0.034 |
| ccng1 | 0.064 | mt-co1 | -0.034 |
| arpc3 | 0.064 | tmsb | -0.034 |
| arhgdia | 0.063 | NC-002333.17 | -0.033 |
| psmb7 | 0.063 | cntn2 | -0.033 |
| pgrmc1 | 0.063 | zfhx3 | -0.033 |
| atp5f1d | 0.063 | cplx2 | -0.032 |
| rap1b | 0.063 | slc18a3a | -0.032 |
| psph | 0.062 | inab | -0.031 |
| cst14a.2 | 0.062 | stx1b | -0.031 |
| tegt | 0.061 | onecut2 | -0.030 |
| cox6b2 | 0.060 | syt2a | -0.030 |
| atp5mc1 | 0.060 | gjd1a | -0.030 |
| si:dkey-204f11.64 | 0.060 | prrt2 | -0.030 |
| gng5 | 0.060 | ndrg4 | -0.030 |
| cox4i1 | 0.060 | tac1 | -0.030 |
| ssr4 | 0.059 | myt1a | -0.029 |
| ndufc2 | 0.059 | kcnc3a | -0.029 |
| cd63 | 0.059 | rbfox1 | -0.029 |