collectin sub-family member 12
ZFIN 
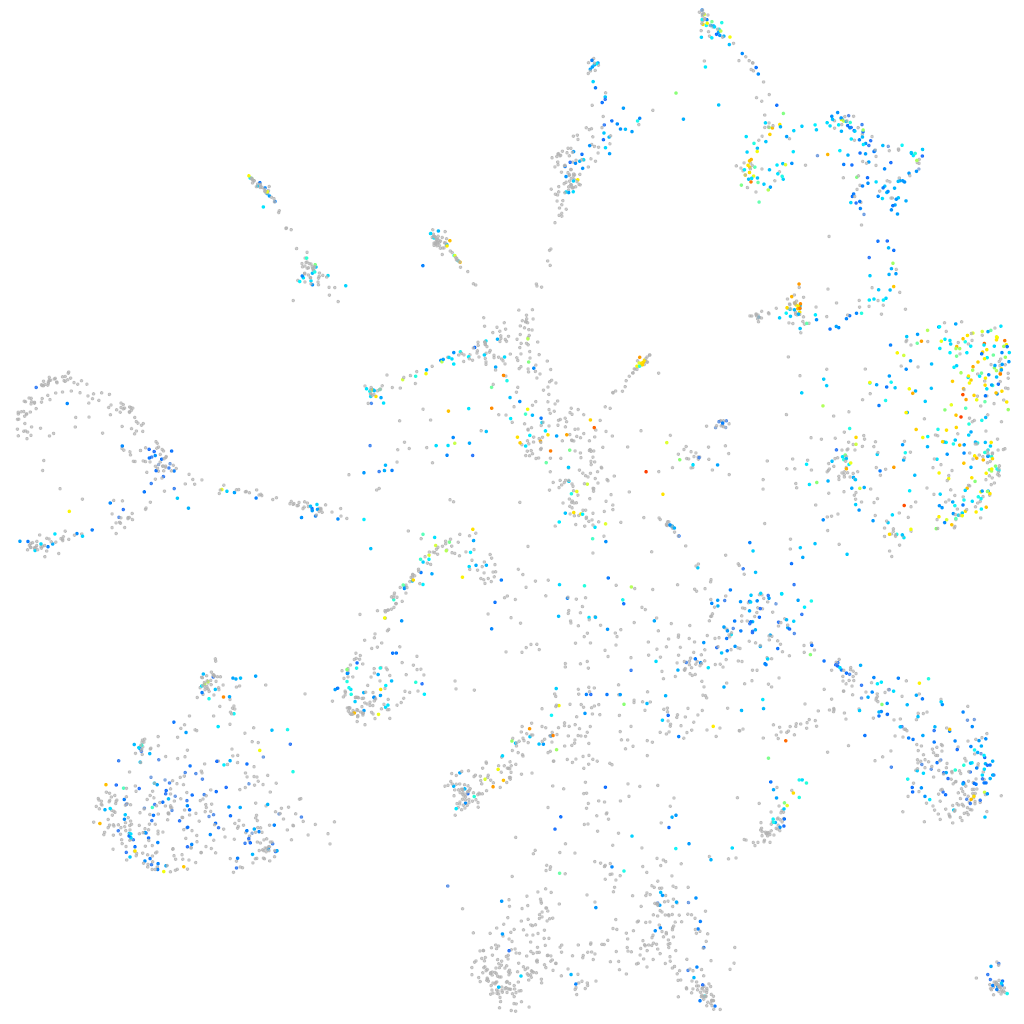
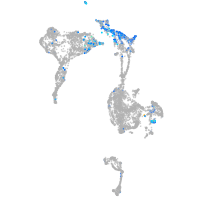
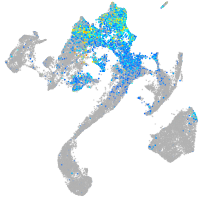
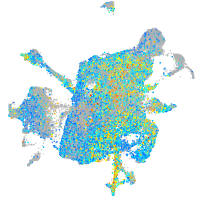
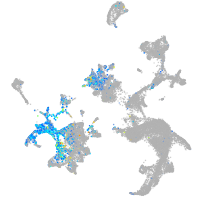
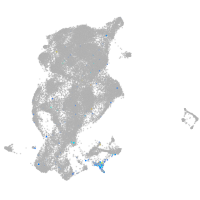
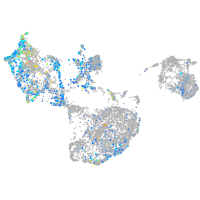
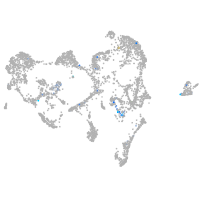
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| podxl | 0.363 | acta2 | -0.234 |
| jam2b | 0.359 | tpm1 | -0.212 |
| aqp1a.1 | 0.357 | tagln | -0.209 |
| krt94 | 0.300 | BX088707.3 | -0.207 |
| bcam | 0.290 | myh11a | -0.206 |
| tmem88b | 0.284 | myl9a | -0.204 |
| cd151 | 0.266 | tpm2 | -0.202 |
| arl4aa | 0.265 | csrp1b | -0.201 |
| col12a1a | 0.263 | cald1b | -0.198 |
| cavin2b | 0.262 | myl6 | -0.194 |
| hspb1 | 0.262 | mylkb | -0.192 |
| cxadr | 0.261 | desmb | -0.186 |
| cavin1b | 0.257 | cnn1b | -0.185 |
| postnb | 0.255 | ptmaa | -0.184 |
| colec11 | 0.251 | lmod1b | -0.179 |
| lrrc15 | 0.244 | ckbb | -0.168 |
| cav1 | 0.244 | rbm24a | -0.149 |
| cd81a | 0.244 | gapdhs | -0.147 |
| bnc2 | 0.243 | fhl1a | -0.146 |
| rhag | 0.238 | pnp4a | -0.142 |
| dag1 | 0.233 | si:ch211-137i24.10 | -0.142 |
| krt8 | 0.230 | gpia | -0.135 |
| krt15 | 0.230 | myocd | -0.134 |
| si:ch211-156j16.1 | 0.228 | rgs2 | -0.134 |
| ehd1b | 0.227 | ak1 | -0.133 |
| ftr82 | 0.224 | BX323087.1 | -0.132 |
| cdh2 | 0.221 | smtna | -0.131 |
| lsp1 | 0.219 | fbxl22 | -0.130 |
| COLEC10 | 0.217 | vcla | -0.130 |
| scarb2a | 0.215 | acta1a | -0.130 |
| cpn1 | 0.213 | tuba8l2 | -0.125 |
| epb41l5 | 0.212 | akap7 | -0.123 |
| nid1a | 0.211 | vdac1 | -0.123 |
| akap12b | 0.209 | acta1b | -0.121 |
| txn | 0.208 | zgc:110699 | -0.120 |