"collagen, type XXI, alpha 1"
ZFIN 
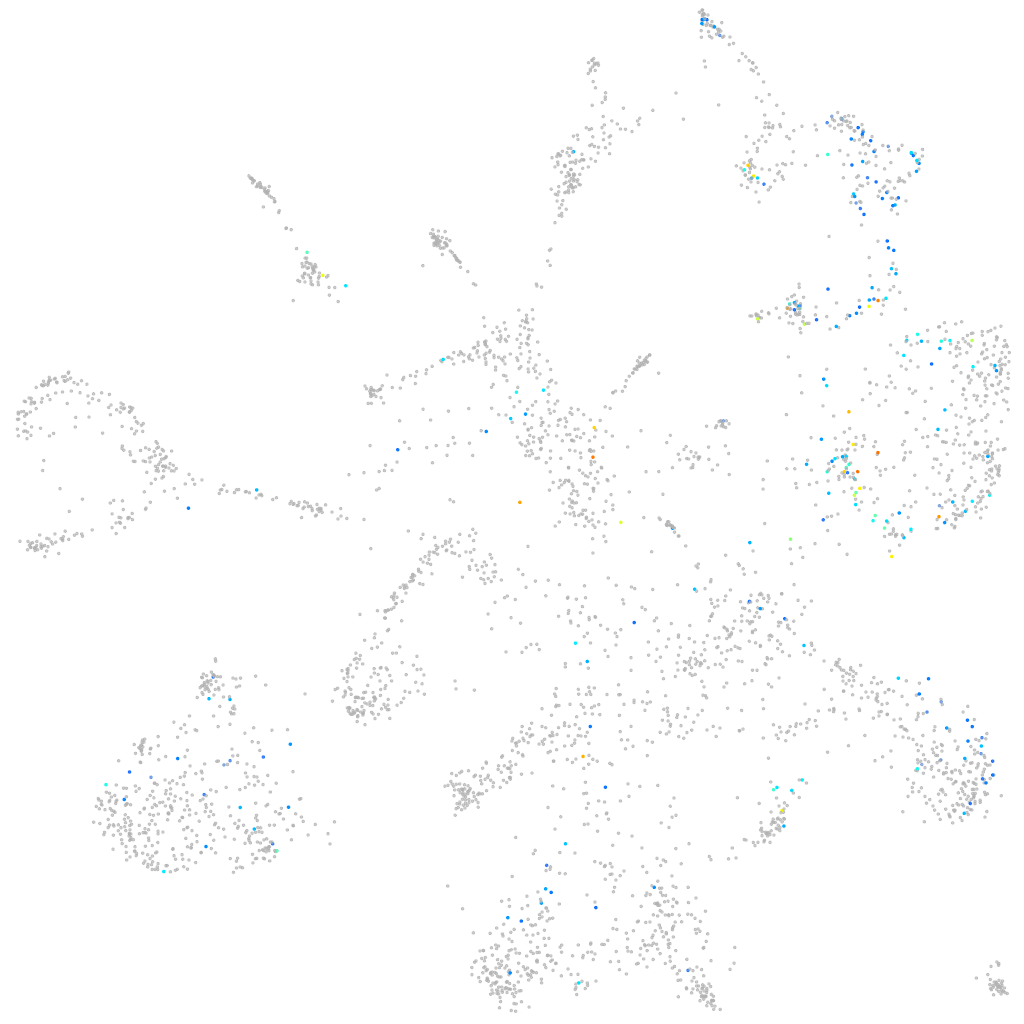
Other cell groups


Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| jam2b | 0.212 | BX088707.3 | -0.134 |
| ftr82 | 0.212 | tpm1 | -0.110 |
| tmem88b | 0.205 | myl9a | -0.109 |
| podxl | 0.196 | acta2 | -0.106 |
| krt94 | 0.179 | tagln | -0.104 |
| wt1b | 0.171 | myh11a | -0.086 |
| fabp11a | 0.168 | myl6 | -0.086 |
| angptl6 | 0.164 | gapdhs | -0.084 |
| cd151 | 0.163 | inka1a | -0.083 |
| COLEC10 | 0.162 | ptmaa | -0.082 |
| tmem88a | 0.160 | mylkb | -0.079 |
| LOC110439708 | 0.153 | ntn5 | -0.077 |
| rspo3 | 0.152 | fhl2a | -0.076 |
| LOC110438146 | 0.150 | tdh | -0.075 |
| cdh2 | 0.149 | ak1 | -0.074 |
| ecm1b | 0.146 | tpi1b | -0.073 |
| nkx3-1 | 0.145 | lmod1b | -0.073 |
| bambib | 0.144 | si:ch211-62a1.3 | -0.073 |
| LOC100535286 | 0.143 | igfbp7 | -0.071 |
| gstm.3 | 0.143 | cox6a1 | -0.070 |
| si:ch211-156j16.1 | 0.141 | ckbb | -0.070 |
| cxcl8b.1 | 0.141 | lims1 | -0.069 |
| cavin2b | 0.141 | cap1 | -0.068 |
| si:ch211-250c4.4 | 0.140 | eno3 | -0.068 |
| bnc2 | 0.138 | tpm2 | -0.066 |
| XLOC-002665 | 0.137 | hmgn3 | -0.065 |
| gata6 | 0.136 | nkx2.3 | -0.065 |
| LOC103911361 | 0.136 | gpia | -0.065 |
| alcamb | 0.135 | myocd | -0.065 |
| cavin1b | 0.134 | cfl1 | -0.064 |
| si:dkey-51d8.1 | 0.133 | atp5pf | -0.064 |
| akap12b | 0.133 | atp5mc1 | -0.064 |
| ankef1b | 0.132 | rbpms2a | -0.064 |
| krt8 | 0.132 | fxyd1 | -0.064 |
| tgm2b | 0.130 | rbm24a | -0.064 |