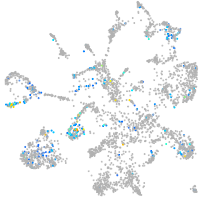
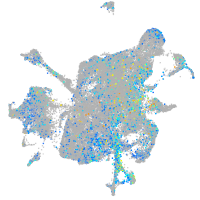
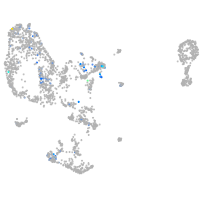
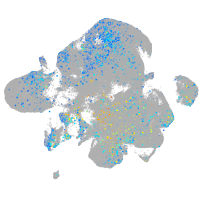
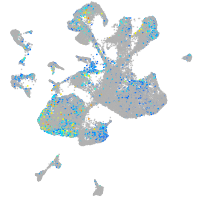
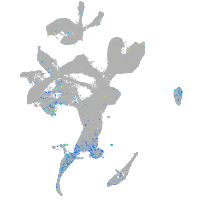
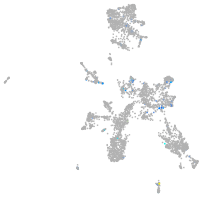
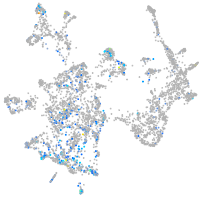
ciliary neurotrophic factor receptor
ZFIN 
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| tpm4a | 0.236 | gapdh | -0.141 |
| bcam | 0.234 | ahcy | -0.127 |
| lsp1 | 0.234 | gamt | -0.119 |
| mcama | 0.212 | eno3 | -0.118 |
| crtap | 0.211 | sod1 | -0.116 |
| pltp | 0.208 | gatm | -0.115 |
| togaram1 | 0.203 | mat1a | -0.110 |
| lamb1a | 0.197 | mdh1aa | -0.110 |
| col8a1a | 0.195 | fbp1b | -0.110 |
| XLOC-042515 | 0.190 | aldob | -0.107 |
| LOC103909937 | 0.190 | gpx4a | -0.107 |
| CABZ01075068.1 | 0.187 | bhmt | -0.106 |
| cd9b | 0.186 | apoa4b.1 | -0.103 |
| hoxb3a | 0.185 | dap | -0.103 |
| col2a1a | 0.184 | scp2a | -0.103 |
| tmem108 | 0.182 | atp5if1b | -0.101 |
| jpt1b | 0.179 | aldh6a1 | -0.101 |
| ndnf | 0.176 | gstt1a | -0.101 |
| cd81a | 0.174 | apoc2 | -0.101 |
| rbms2b | 0.173 | glud1b | -0.100 |
| qkia | 0.173 | apoa1b | -0.098 |
| emid1 | 0.172 | abat | -0.098 |
| gdf6a | 0.172 | agxtb | -0.096 |
| fermt1 | 0.172 | tpi1b | -0.096 |
| fgfrl1b | 0.171 | suclg1 | -0.095 |
| zfp36l1a | 0.170 | sod2 | -0.095 |
| fstl1a | 0.170 | suclg2 | -0.095 |
| si:dkey-27b3.4 | 0.169 | pklr | -0.093 |
| nav3 | 0.168 | pgk1 | -0.093 |
| si:ch211-264f5.8 | 0.165 | grhprb | -0.093 |
| BX469925.3 | 0.164 | g6pca.2 | -0.093 |
| txnipa | 0.164 | gnmt | -0.092 |
| cnn2 | 0.162 | haao | -0.091 |
| hmga2 | 0.161 | pnp4b | -0.091 |
| lin28a | 0.161 | sult2st2 | -0.090 |