canopy FGF signaling regulator 4
ZFIN 
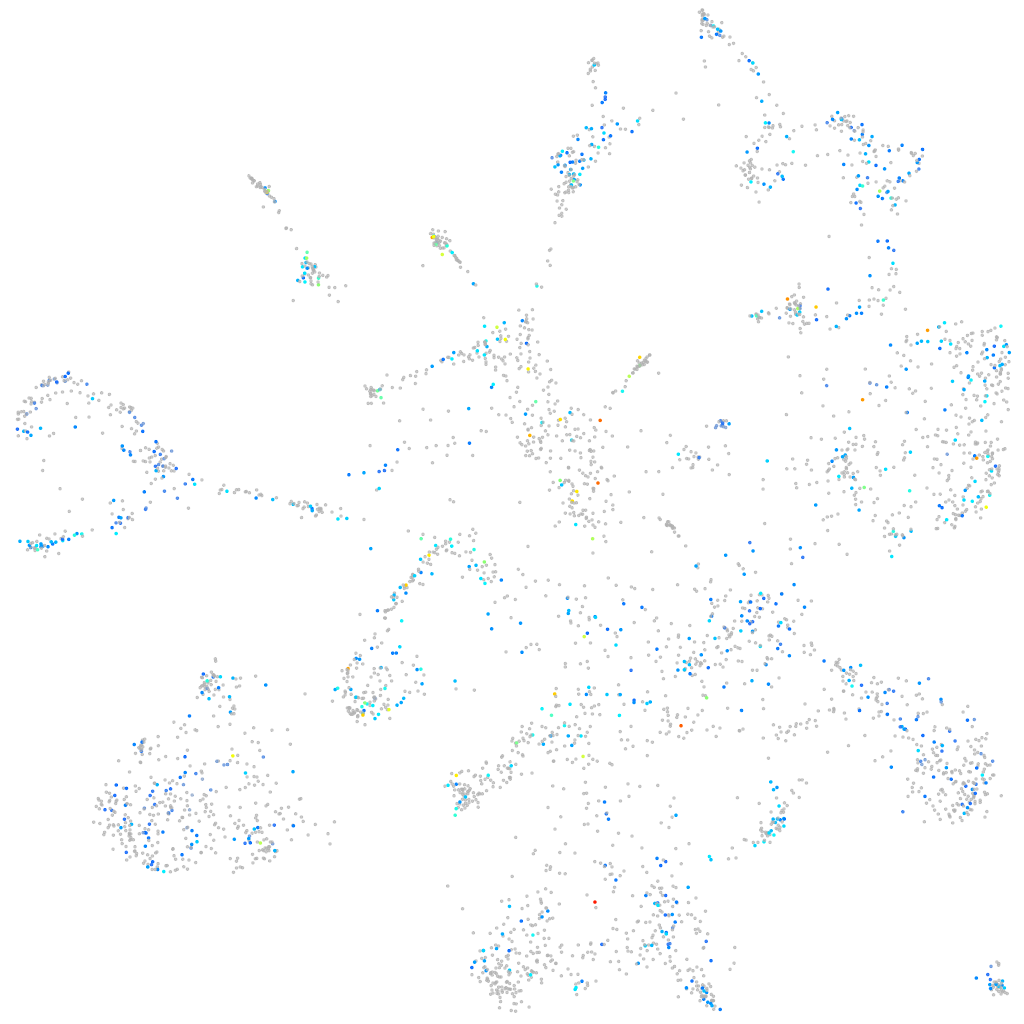
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| ssr4 | 0.149 | desmb | -0.061 |
| fkbp7 | 0.145 | basp1 | -0.058 |
| ckap4 | 0.141 | ckbb | -0.057 |
| tmed2 | 0.140 | XLOC-025423 | -0.056 |
| ssr3 | 0.138 | csrp1b | -0.056 |
| fkbp14 | 0.136 | cald1b | -0.056 |
| ddost | 0.129 | XLOC-005448 | -0.052 |
| sparc | 0.127 | arhgap20b | -0.052 |
| pdia3 | 0.127 | zgc:158463 | -0.051 |
| pdia6 | 0.123 | smtnb | -0.051 |
| crtap | 0.121 | vcla | -0.050 |
| hspa5 | 0.120 | kif11 | -0.049 |
| ssr2 | 0.120 | BX322618.1 | -0.048 |
| sec11a | 0.119 | rgs2 | -0.048 |
| selenof | 0.117 | cnn1b | -0.048 |
| nucb2a | 0.116 | fras1 | -0.047 |
| xbp1 | 0.116 | myofl | -0.047 |
| dad1 | 0.116 | dlb | -0.047 |
| sec61b | 0.115 | itpk1b | -0.046 |
| ppib | 0.114 | cpa5 | -0.045 |
| calr | 0.113 | rtn1a | -0.045 |
| tspan7 | 0.113 | ctrb1 | -0.045 |
| XLOC-035413 | 0.112 | gapdh | -0.045 |
| tram1 | 0.111 | pdlim3b | -0.045 |
| lman1 | 0.111 | CR753876.1 | -0.044 |
| kdelr2b | 0.111 | prss59.1 | -0.044 |
| kdelr3 | 0.111 | slitrk6 | -0.044 |
| myl12.1 | 0.110 | tagln | -0.044 |
| manf | 0.110 | actc1b | -0.044 |
| ostc | 0.109 | nova1 | -0.044 |
| calua | 0.107 | CELA1 (1 of many) | -0.043 |
| mfap2 | 0.106 | LOC110439372 | -0.043 |
| prdx4 | 0.106 | si:ch211-196c10.15 | -0.043 |
| tmed9 | 0.106 | si:ch211-137i24.10 | -0.043 |
| ucp2 | 0.106 | anln | -0.043 |